Machine learning can use patients’ demographic information and previous clinical history to help physicians select the antibiotics most likely to successfully treat urinary tract infections, despite growing levels of resistance.
Recent years have seen a worrying increase in the levels of antibiotic resistance of many bacterial infections. Antibiotic resistance not only makes treating bacterial infections difficult but also decreases the effectiveness of antibiotic prophylaxis needed for safe surgeries, organ transplantation and cancer treatment1. There is an urgent need for new effective antibiotics; however, the antibiotic-development pipeline is dry. Without government intervention, research to develop new antibiotics is rarely profitable, and consequently most major pharmaceutical companies have left the field1. Therefore, using the antibiotics that we have at our disposal in an optimized way is crucial, to avoid the risks of both treatment failure and further increasing resistance levels2. In this issue of Nature Medicine, Yelin et al.3 describe a strategy for combating the drug resistance caused by mismatched antibiotic prescriptions in urinary tract infections (UTIs).
Antibiotic treatment for various bacterial infections, such as UTIs, is typically started empirically without knowledge of the antibiotics to which the bacteria causing the infection are susceptible. UTIs are one of the most common infections encountered in primary care4 and account for 29–66% of antibiotic prescriptions in care-home settings5. The bacteria causing UTIs are often carried asymptomatically in the human body and are therefore frequently exposed to antibiotics, including those taken to treat other infections6. Consequently, the bacteria causing UTIs are frequently resistant to various commonly used antibiotics7. Physicians treating patients with UTIs are therefore routinely faced with several difficult questions. Which antibiotic is most likely to cure the patient? How can the usefulness of antibiotics be preserved in the long term? Should published guidelines be followed, or is there something special about the patient that justifies a different course of action?
Yelin et al.3 analyzed data from more than 700,000 UTIs occurring in ~315,000 patients between 2007 and 2017 in Israel, including information on the patients’ demographics, clinical history and previous use of antibiotics. The resistance of cultured bacterial pathogens from the UTIs against six antibiotics frequently prescribed against UTIs was found to be associated with several demographic factors, especially the patient age, sex and residence in a retirement home. Many patients contracted multiple UTIs during the 10-year study period, in which case, the resistance profiles were often similar among their UTIs, thus suggesting either relapse or reinfection from the same source, depending on the time span. The history of antibiotic use (as measured on the basis of information on antibiotic purchase) was also found to be associated with antimicrobial resistance of the bacterial pathogen, and the strongest association was found between use and resistance of the same antibiotic; however, other less expected associations also existed, as previously noted6. There are various mechanisms by which the use of one antibiotic might select for resistance against another unrelated antibiotic3,6,8.
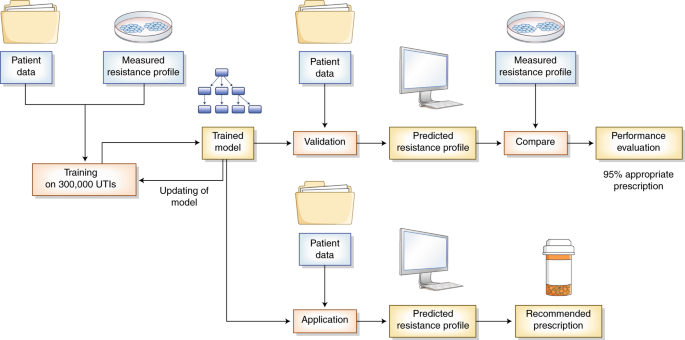
Beyond its importance in understanding the drivers of antimicrobial resistance, both within hosts and at the population scale8,9, the study by Yelin et al.3 has the potential for application to personalized medicine. The authors used a machine-learning method, called gradient-boosting decision trees, to derive an algorithm that takes as input all available information on demographic factors, previous infection history and antibiotic use (as measured by antibiotic purchase), and returns as output a prediction of the resistance profile of a new infection. In machine learning, algorithms are trained to perform complex tasks by recognizing patterns in large high-dimensional datasets. In contrast to more traditional statistical regression techniques, no explicit relationships are required to be assumed between the input and output parameters. Because part of the recognized patterns might be due to random variation and might be specific to the training dataset, evaluating the performance of the trained model in a test dataset is crucial (Fig. 1).
Yelin et al. used data from more than 300,000 UTIs, including previous antibiotic use, to develop an algorithm that prescribed an incorrect antibiotic 5% of the time, as compared with 9% of the time by physicians. The machine-learning method is first trained on a large number of patient records and antibiotic-resistance measurements. The resulting trained model is able to predict resistance profiles on the basis of patient data, and the results in turn are used to select the antibiotic prescription most likely to succeed in curing the infection. The performance of this trained model is evaluated on a separate set of patients to allow for comparison between the predicted and observed resistance measurements.
In this study, to fairly assess the accuracy of the predictions, the model was trained with only the first 9 years of data, so that the remaining tenth year could be used for independent testing. This benchmark demonstrated that the model has a strong capacity to predict resistance to specific antibiotics. Regarding the choice among the six drugs frequently prescribed against UTIs, in 8.5% of cases, the physicians prescribed an inappropriate drug, that is, one to which the infection was resistant. This rate is only slightly better than the 10% of inappropriate prescriptions that would have occurred if the drug were chosen at random. However, when the predictive computer model was used, the proportion of inappropriate prescription decreased to 5%.
The study by Yelin et al.3 paves the way for the use of machine-learning-assisted decision-making in prescribing antibiotics against UTIs in a way that minimizes the risk of treatment failure. However, the model would have to be retrained separately on relevant data for application to other populations and to keep up with current trends in resistance. The availability of such training data may be limited in some settings, such as in patient populations in which doctors do not routinely send samples for antibiotic-susceptibility testing. In addition, detailed data about patients’ history of antibiotic use and previous test results are not always available to the prescribing physicians. It would be interesting to determine in future work whether a model with less detailed input-data requirements could also improve the selection of antibiotics that are effective against UTIs. When detailed data are available for most patients, similar strategies could be used for the treatment of other bacterial infections, as has recently been proposed for bloodstream infections in a hospital setting10.
Accurate prediction of resistance against different antibiotics is directly beneficial from the patient point of view, because it avoids treatment failures. Such prediction could have additional long-term benefits, for example, enabling the use of more targeted antibiotics, decreasing the need to use multiple antibiotics to cure the same infection and lowering the risk of onward transmission2,11. All these factors would contribute to a decrease in the selective advantage of resistant pathogens, and, because resistance often has a substantial fitness cost, could consequently decrease the overall resistance levels and ease the global threat posed by antibiotic resistance12.
(원문: 여기를 클릭하세요~)