Abstract
Nature uses 64 codons to encode the synthesis of proteins from the genome, and chooses 1 sense codon—out of up to 6 synonyms—to encode each amino acid. Synonymous codon choice has diverse and important roles, and many synonymous substitutions are detrimental. Here we demonstrate that the number of codons used to encode the canonical amino acids can be reduced, through the genome-wide substitution of target codons by defined synonyms. We create a variant of Escherichia coli with a four-megabase synthetic genome through a high-fidelity convergent total synthesis. Our synthetic genome implements a defined recoding and refactoring scheme—with simple corrections at just seven positions—to replace every known occurrence of two sense codons and a stop codon in the genome. Thus, we recode 18,214 codons to create an organism with a 61-codon genome; this organism uses 59 codons to encode the 20 amino acids, and enables the deletion of a previously essential transfer RNA.
(원문: 여기를 클릭하세요~)
Construction of an Escherichia coli genome with fewer codons sets records
The biggest synthetic genome so far has been made, with a smaller set of amino-acid-encoding codons than usual — raising the prospect of encoding proteins that contain unnatural amino-acid residues.
Over the past decade, decreases in the costs of chemically synthesizing DNA and improved methods for assembling DNA fragments have enabled researchers to scale up synthetic biology to the level of generating entire chromosomes and genomes. So far, synthetic DNA has been constructed with up to one million base pairs, notably a set of chromosomes from the yeast Saccharomyces cerevisiae and several versions of the genome of the bacterium Mycoplasma mycoides1,2. Now, writing in Nature, Fredens et al.3 report the completion of a 4-million-base-pair synthetic version of the Escherichia coli genome. This is a landmark in the emerging field of synthetic genomics, and finally applies the technology to the laboratory’s workhorse bacterium.
Synthetic genomics offers a new way of understanding the rules of life, while at the same time moving synthetic biology towards a future in which genomes can be written to design. The pioneers in the field — the researchers at the J. Craig Venter Institute in Rockville, Maryland — have used this method to better define the minimal set of genes required for a free-living cell. By adopting an approach that involves redesigning genome segments by computer, chemically synthesizing the fragments and then assembling them, these pioneers succeeded2 in reducing the size of the M. mycoidesgenome by around 50%. Doing the same with just genome-editing tools would be much more laborious, as past work with E. coli demonstrates: here, gene-deletion methods have removed, at best, only 15% of the genome4.
Fredens and colleagues used this reduced genome from E. coli as the template for a synthetic genome with another kind of minimization in mind — codon reduction. The genetic code has inherent redundancy: there are 64 codons (triplets of ‘letters’, or bases) to encode just 20 amino acids plus the ‘start’ and ‘stop’ points that mark the beginning and end of a stretch of protein-coding sequence. This redundancy means, for example, that there are six codons that encode the amino acid serine, and three possible stop codons. Through design, synthesis and assembly, Fredens et al.3 have been able to construct an E. coli genome that uses only 61 of the 64 available codons in its protein-coding sequences, replacing two serine codons and one stop codon with synonyms (codons that are ‘spelt’ differently but give the same instruction). Past work using genome-editing tools has already produced a synthetic E. coli that uses just 63 of the 64 codons, but this required only the stop codons with the sequence TAG (of which there were just 321 around the genome) to be changed to an alternative stop codon5. Reduction to 61 codons demanded that a whopping 18,214 codons be changed, necessitating a genome-synthesis approach.
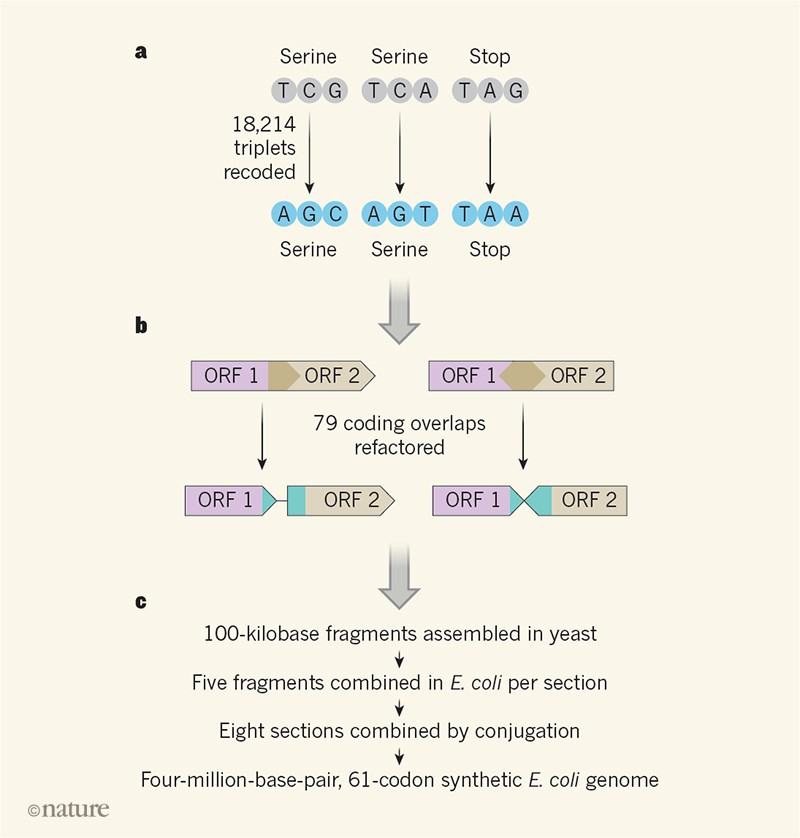
Fredens and colleagues built their synthetic E. coli genome by using large-scale DNA-assembly and genome-integration methods that they had developed previously6 to probe the limits of codon changes in E. coli. In their approach (Fig. 1), DNA is computationally designed, chemically synthesized and assembled in 100-kilobase fragments in vectors in S. cerevisiae; these vectors are then taken up by E. coli and integrated into the genome in the direct place of the equivalent natural region. Iterating this process five times resulted in 500-kilobase sections of DNA being replaced by synthetic versions. Eight strains of E. coli were produced in this way, each harbouring synthetic DNA sections that covered a different region of the genome. These sections were then combined using conjugation methods to make the complete synthetic genome.
Figure 1 | Design and construction of a recoded genome. a, Fredens et al.3 recoded three base triplets (codons) — TCG and TCA, which encode the amino acid serine, and TAG, a stop codon that marks the end of a protein-coding sequence — to alternatives that have the same functions (AGC, AGT and TAA respectively) in the genome of the bacterium Escherichia coli. b, In some genomic locations, open reading frames (ORFs; protein-coding regions) overlap, and a change in the codons of one ORF might produce an unwanted change in the overlapping region. Fredens et al. ‘refactored’ these ORFs to separate them, as illustrated for ORF1 and ORF2 (the two ORFs on the left are ‘read’ in the same direction; the two on the right are read in opposite directions). c, Redesigned DNA was synthesized and assembled into 100-kilobase fragments in the yeast Saccharomyces cerevisiae; fragments were then combined into sections and integrated into the E. coligenome. The sections were brought together to generate the complete functional synthetic genome.
The large-scale construction was impressively successful, with very low off-target mutation rates, but was not without its challenges. Many genes in the E. coli genome partially overlap with others, and in 91 cases the overlapping regions contained codons that needed to be changed. This is complex because synonymous alterations in one protein-coding sequence might alter the amino acids encoded by the overlapping one. To tackle this, the team ‘refactored’ 79 locations in the genome, duplicating the sequence to separate out overlapped coding sequences into individual recoded ones (Fig. 1). Although this approach was generally successful, it did require careful debugging in a few cases in which refactoring also altered gene regulation.
The final strain proved viable and was able to grow in a range of typical laboratory conditions, albeit a little less vigorously than its natural counterpart. It no longer uses the stop codon TAG or the two serine codons TCG and TCA, so the cellular machinery that recognizes these can now be either deleted or reassigned to recruit ‘non-canonical’ amino acids beyond the usual 20 used by most living cells. Such recruitment has already been shown to be useful in the 63-codon E. coli, both for biotechnology projects, in which non-canonical amino acids are encoded into desired sequence positions to provide residues that can take part in chemical reactions that natural proteins can’t; and for biosafety reasons, in that the natural transfer of readable DNA-encoded information in and out of the synthetic E. coli is limited because the cell operates with a slightly different genetic code from the rest of the natural world5. Expect all of these applications to be expanded in the new 61-codon E. coli, which has the potential to encode the use of more than one non-canonical amino acid, and to generate a more stringent genetic firewall (because 3 of the 64 codons are no longer recognized).
Synthesis of a 4-million-base-pair genome and reduction of the genetic code to 61 codons are new records for synthetic genomics, but might not be for much longer. The international Sc2.0 consortium is closing in on synthesizing all 16 chromosomes of the 12-million-base-pair S. cerevisiae genome — the first synthetic genome of a eukaryotic organism, the group that includes plants, animals and fungi — and the synthesis of a 57-codon E. coli genome is also under way1,7. A genome of the bacterium Salmonella Typhimurium that has two fewer codons than the natural organism is also being constructed8. This could one day enable bacteria with synthetic genomes to be used as cell-based technologies in the human gut.
From a technological standpoint, the most interesting aspect of all these different projects is that the workflows for synthetic-genome construction are remarkably similar, with kilobase sections of synthesized DNA being assembled (by the process of homologous recombination) into 50- to 100-kilobase pieces in yeast cells, and these pieces then being used to replace natural sequences inside the target organism (by selectable recombination methods). Standardization of methods will enable steps to be automated and more research groups to enter the field. Genome minimization and codon reduction are just the first uses of this new technology, which could one day give us functionally reorganized genomes and genomes that are custom designed to direct cells to perform specialized tasks.
Nature 569, 492-494 (2019)
(원문: 여기를 클릭하세요~)
Scientists Created Bacteria With a Synthetic Genome. Is This Artificial Life?
In a milestone for synthetic biology, colonies of E. coli thrive with DNA constructed from scratch by humans, not nature.

Scientists have created a living organism whose DNA is entirely human-made — perhaps a new form of life, experts said, and a milestone in the field of synthetic biology.
Researchers at the Medical Research Council Laboratory of Molecular Biology in Britain reported on Wednesday that they had rewritten the DNA of the bacteria Escherichia coli, fashioning a synthetic genome four times larger and far more complex than any previously created.
The bacteria are alive, though unusually shaped and reproducing slowly. But their cells operate according to a new set of biological rules, producing familiar proteins with a reconstructed genetic code.
The achievement one day may lead to organisms that produce novel medicines or other valuable molecules, as living factories. These synthetic bacteria also may offer clues as to how the genetic code arose in the early history of life.
“It’s a landmark,” said Tom Ellis, director of the Center for Synthetic Biology at Imperial College London, who was not involved in the new study. “No one’s done anything like it in terms of size or in terms of number of changes before.”
Each gene in a living genome is detailed in an alphabet of four bases, molecules called adenine, thymine, guanine and cytosine (often described only by their first letters: A, T, G, C). A gene may be made of thousands of bases.
Genes direct cells to choose among 20 amino acids, the building blocks of proteins, the workhorses of every cell. Proteins carry out a vast number of jobs in the body, from ferrying oxygen in the blood to generating force in our muscles.
Nine years ago, researchers built a synthetic genome that was one million base pairs long. The new E. coli genome, reported in the journal Nature, is four million base pairs long and had to be constructed with entirely new methods.
The new study was led by Jason Chin, a molecular biologist at the M.R.C. laboratory, who wanted to understand why all living things encode genetic information in the same baffling way.
The production of each amino acid in the cell is directed by three bases arranged in the DNA strand. Each of these trios is known as a codon. The codon TCT, for example, ensures that an amino acid called serine is attached to the end of a new protein.
Since there are only 20 amino acids, you’d think the genome only needs 20 codons to make them. But the genetic code is full of redundancies, for reasons that no one understands.
Amino acids are encoded by 61 codons, not 20. Production of serine, for example, is governed by six different codons. (Another three codons are called stop codons; they tell DNA where to stop construction of an amino acid.)
Like many scientists, Dr. Chin was intrigued by all this duplication. Were all these chunks of DNA essential to life?
“Because life universally uses 64 codons, we really didn’t have an answer,” Dr. Chin said. So he set out to create an organism that could shed some light on the question.
 A colored scanning electronic micrograph of synthesized Mycoplasma mycoides, bacteria with a genome containing one million base pairs. Now scientists have created an E.coli genome four times larger. CreditThomas Deerinck, NCMIR/Science Source
A colored scanning electronic micrograph of synthesized Mycoplasma mycoides, bacteria with a genome containing one million base pairs. Now scientists have created an E.coli genome four times larger. CreditThomas Deerinck, NCMIR/Science Source
After some preliminary experiments, he and his colleagues designed a modified version of the E. coli genome on a computer that only required 61 codons to produce all of the amino acids the organism needs.
Instead of requiring six codons to make serine, this genome used just four. It had two stop codons, not three. In effect, the researchers treated E. coli DNA as if it were a gigantic text file, performing a search-and-replace function at over 18,000 spots.
Now the researchers had a blueprint for a new genome four million base pairs long. They could synthesize the DNA in a lab, but introducing it into the bacteria — essentially substituting synthetic genes for those made by evolution — was a daunting challenge.
The genome was too long and too complicated to force into a cell in one attempt. Instead, the researchers built small segments and swapped them piece by piece into E. coli genomes. By the time they were done, no natural segments remained.
Much to their relief, the altered E. coli did not die. The bacteria grow more slowly than regular E. coli and develop longer, rod-shaped cells. But they are very much alive.
Dr. Chin hopes to build on this experiment by removing more codons and compressing the genetic code even further. He wants to see just how streamlined the genetic code can be while still supporting life.
The Cambridge team is just one of many racing in recent years to build synthetic genomes. The list of potential uses is a long one. One attractive possibility: Viruses may not be able to invade recoded cells.
Many companies today use genetically engineered microbes to make medicines like insulin or useful chemicals like detergent enzymes. If a viral outbreak hits the fermentation tanks, the results can be catastrophic. A microbe with synthetic DNA might be made immune to such attacks.
Recoding DNA could also allow scientists to program engineered cells so that their genes won’t work if they escape into other species. “It creates a genetic firewall,” said Finn Stirling, a synthetic biologist at Harvard Medical School who was not involved in the new study.
Researchers are also interested in recoding life because it opens up the opportunity to make molecules with entirely new kinds of chemistry.
Beyond the 20 amino acids used by all living things, there are hundreds of other kinds. A compressed genetic code will free up codons that scientists can use to encode these new building blocks, making new proteins that carry out new tasks in the body.
James Kuo, a postdoctoral researcher at Harvard Medical School, offered a note of caution. Tacking bases together to make genomes remains enormously costly.
“It’s just way too expensive for academic groups to keep pursuing,” Dr. Kuo said.
But E. coli is a workhorse of laboratory research, and now it’s clear that its genome can be synthesized. It’s not hard to imagine that prices will fall as demands for custom, synthetic DNA rise. Researchers could apply Dr. Chin’s methods to yeast or other species.
“In theory, you could recode anything,” said Mr. Stirling.
(원문: 여기를 클릭하세요~)