“뜻밖의” 외계생명체 가능 입증…탐색 방식 재검토 필요성 제기돼
새로 합성된 ‘하치모지 DNA‘기존 4개 요소(적색·녹색·청색·황색 )에다 새로 4개(분홍색, 보라색, 오렌지색, 청록색)가 추가됐다. [인디애나 의과대학원 제공]
과학자들이 디옥시리보핵산(DNA)처럼 유전 정보를 저장하고 전달할 수 있는 분자시스템을 합성해 냈다. 이는 새로운 생명체 형태는 아니지만 DNA에 기반한 지구 생명체와는 전혀 다른 외계 생명체가 존재할 수 있다는 점을 보여주는 것이다.
미국항공우주국(NASA)은 NASA의 연구비 지원을 받아 이뤄진 이번 연구결과가 외계 생명체 탐색 방식에 대한 재검토 필요성을 나타내는 것으로 받아들이고 있다.
NASA에 따르면 미국 ‘응용 분자진화 재단(FfAME)의 스티븐 베너 박사가 이끄는 연구팀은 아데닌(A)과 시토신(S), 구아닌(G), 티민(T) 등 DNA를 구성하는 4개 염기에다 이를 모방한 다른 4개 요소를 추가한 분자 정보시스템을 합성하는 데 성공했다고 국제학술지 ‘사이언스(Science)’ 최신호에 밝혔다.
DNA는 지구상의 모든 생명체가 유전정보를 저장하고 다음 세대에 전달하는 복합분자로 A, S, G, T 염기 순서 조합을 이용하는데 베너 박사 연구팀은 이를 8개로 늘려 새로운 분자시스템을 합성했다.
이 분자시스템은 기존 DNA와 마찬가지 형태의 이중나선 구조로 유전 정보를 저장하고 전달하는 것으로 나타났다.
연구팀은 이 분자시스템에 ‘8개 문자’를 뜻하는 일본어를 이용해 ‘하치모지 DNA‘라는 이름을 붙였다.
베너 박사는 “하치모지 DNA의 형태와 크기, 구조 등의 역할을 면밀히 분석함으로써 외계 생명체의 유전 정보를 저장하고 있을 수도 있는 분자 구조의 형태에 대한 이해를 넓히게 됐다”고 했다.
NASA는 과학자들이 외계 생명체의 존재와 유전자 시스템에 관한 답을 얻기까지 넘어야 할 산이 많지만 이번 연구 결과는 지구 기준으로 생명체가 살 수 없을 것으로 여겨지는 환경에서 전혀 상상할 수 없는 생명체가 살 수 있는 방식을 연구하는 새로운 계기가 될 것이라고 지적했다.

외계생명체 탐색의 1순위 기준은?[M.네베우/NASA 우주생물학프로그램/UCLA/JPL–Caltech 제공]
예컨대 목성의 위성인 유로파나 토성의 위성인 엔켈라두스는 두꺼운 얼음층 밑에 바다가 있는 것으로 알려져 있는데 지구 DNA와 전혀 다른 형태의 분자시스템을 가진 생명체가 있다면 우리가 알아보지 못할 수도 있으며, 새 분자시스템 합성이 이런 문제를 푸는 연구의 출발점이라는 것이다.
NASA 우주생물학 담당 수석과학자인 메리 보이텍은 “(외계 생명체 탐색) 장비 설계와 임무 개념이 가능한 존재에 대한 더 확대된 이해를 토대로 한다면 포괄적이고 효과적인 탐색으로 이어질 것”이라고 했다.
NASA 행성과학부문 책임자인 로리 글레이즈도 “생명체 탐사가 행성과학 임무에서 점점 더 중요한 부분이 돼가고 있으며, 이번 연구결과는 효과적인 장비와 실험을 개발해 우리가 살펴보는 범위를 확대할 수 있게 해줄 것”이라고 밝혔다.
(원문: 여기를 클릭하세요~)
.
Synthetic DNA seems to behave like the natural variety, suggesting that a broader swathe of chemicals could support life than the four that evolved on Earth.
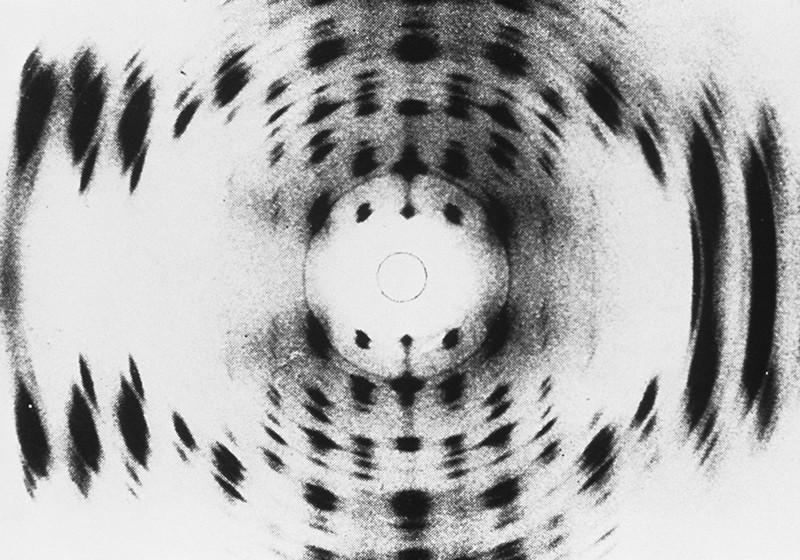
An X-ray diffraction image of part of a molecule of DNA. The new, 8-letter version, is similarly stable. Credit: Science Source/Science Photo Library
The DNA of life on Earth naturally stores its information in just four key chemicals — guanine, cytosine, adenine and thymine, commonly referred to as G, C, A and T, respectively.
Now scientists have doubled this number of life’s building blocks, creating for the first time a synthetic, eight-letter genetic language that seems to store and transcribe information just like natural DNA.
In a study published on 22 February in Science1, a consortium of researchers led by Steven Benner, founder of the Foundation for Applied Molecular Evolution in Alachua, Florida, suggests that an expanded genetic alphabet could, in theory, also support life.
“It’s a real landmark,” says Floyd Romesberg, a chemical biologist at the Scripps Research Institute in La Jolla, California. The study implies that there is nothing particularly “magic” or special about those four chemicals that evolved on Earth, says Romesberg. “That’s a conceptual breakthrough,” he adds.
Normally, as a pair of DNA strands twist around each other in a double helix, the chemicals on each strand pair up: A bonds to T, and C bonds with G.
For a long time, scientists have tried to add more pairs of these chemicals, also known as bases, to this genetic code. For example, Benner first created ‘unnatural’ bases in the 1980s. Other groups have followed, with Romesberg’s lab making headlines in 2014 after inserting a pair of unnatural bases into a living cell.
But the latest study is the first to systematically demonstrate that the complementary unnatural bases recognise and bind to each other, and that the double helix that they form holds its structure.
Benner’s team, which includes researchers from various US companies and institutions, created the synthetic letters by tweaking the molecular structure of the regular bases. The letters of DNA pair up because they form hydrogen bonds: each contains hydrogen atoms, which are attracted to nitrogen or oxygen atoms in their partner. Benner explains that it’s a bit like Lego bricks that snap together when the holes and prongs line up.
By adjusting these holes and prongs, the team has come up with several new pairs of bases, including a pair named S and B, and another called P and Z2. In the latest paper, they describe how they combine these four synthetic bases with the natural ones. The researchers call the resulting eight-letter language ‘hachimoji’ after the Japanese words for ‘eight’ and ‘letter’. The additional bases are each similar in shape to one of the natural four, but have variations in their bonding patterns.
The researchers then conducted a series of experiments that showed that their synthetic sequences shares properties with natural DNA that are essential for supporting life.
Data retrieval
To work as an information storage system, DNA has to follow predictable rules, so the team first demonstrated that, in a similar way to regular bases, the synthetic bases reliably formed pairs. They created hundreds of molecules of the synthetic DNA and found that the letters bound to their partners predictably.
They then showed that the structure of the double helices remained stable no matter what order the synthetic bases were in. This is important because for life to evolve, DNA sequences need to be able to vary without the whole structure falling apart. Using X-ray diffraction, the team showed that three different sequences of the synthetic DNA retained the same structure when crystallised.
This is a substantial advance, says Philipp Holliger, a synthetic biologist at the MRC Laboratory of Molecular Biology in Cambridge, UK, because other methods of expanding the genetic alphabet are not as structurally sound. Instead of chemicals that use hydrogen bonds to pair up, these other approaches use water-repelling molecules as their bases. These can be placed at intervals in-between the natural letters, but the structure of DNA breaks down if they are placed in a row.
Finally, the team showed that the synthetic DNA could be faithfully transcribed into RNA. “The ability to store information is not very interesting for evolution,” says Benner. “You have to be able to transfer that information into a molecule that does something.”
Converting DNA into RNA is a key step for translating genetic information into proteins, the workhorses of life. But some RNA sequences, known as aptamers, can themselves bind to specific molecules.
Benner’s team created synthetic DNA that codes for a certain aptamer and then confirmed that the transcription had occurred and the RNA sequence functioned correctly.
Holliger says that the work is an exciting starting point, but there is still a substantial distance to go before reaching a true eight-letter synthetic genetic system. One key question, for example, will be whether the synthetic DNA can be replicated by polymerases, the enzymes responsible for synthesizing DNA inside organisms during cell division. This has been demonstrated for other methods such as Romesberg’s, which uses water-repelling bases.
Variety of life
Still, Benner says that the work shows that life could potentially be supported by DNA bases with different structures from the four that we know, which could be relevant in the search for signatures of life elsewhere in the Universe.
Adding letters to DNA could also have more down-to-earth applications.
With more diversity in the genetic building blocks, scientists could potentially create RNA or DNA sequences that can do things better than the standard four letters, including functions beyond genetic storage.
For example, Benner’s group previously showed that strands of DNA that included Z and P were better at binding to cancer cells than sequences with just the standard four bases3. And Benner has set up a company which commercialises synthetic DNA for use in medical diagnostics.
The researchers could potentially use their synthetic DNA to create novel proteins as well as RNA. Benner’s team has also developed further pairs of new bases, opening up the possibility of creating DNA structures that contain 10 or even 12 letters. But the fact that the researchers have already expanded the genetic alphabet to eight is in itself remarkable, says Romesberg. “It’s already doubling what nature has.”
(원문: 여기를 클릭하세요~)