Abstract
Much of the functionality of multicellular systems arises from the spatial organization and dynamic behaviours within and between cells. Current single-cell genomic methods only provide a transcriptional ‘snapshot’ of individual cells. The real-time analysis and perturbation of living cells would generate a step change in single-cell analysis. Here we describe minimally invasive nanotweezers that can be spatially controlled to extract samples from living cells with single-molecule precision. They consist of two closely spaced electrodes with gaps as small as 10–20 nm, which can be used for the dielectrophoretic trapping of DNA and proteins. Aside from trapping single molecules, we also extract nucleic acids for gene expression analysis from living cells without affecting their viability. Finally, we report on the trapping and extraction of a single mitochondrion. This work bridges the gap between single-molecule/organelle manipulation and cell biology and can ultimately enable a better understanding of living cells.
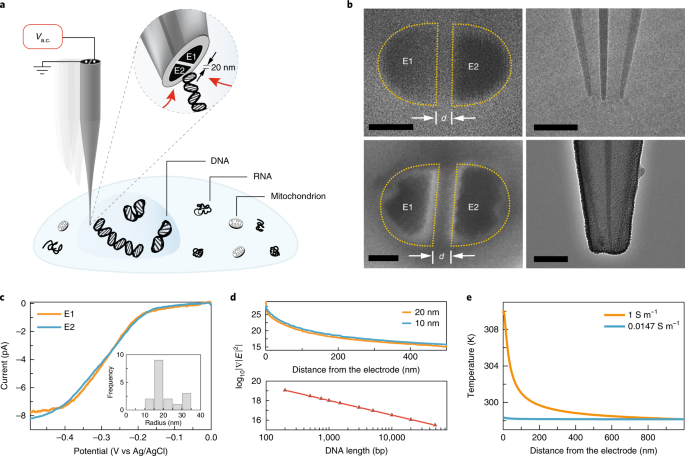
a, Application of an a.c. voltage on the nanotweezer generates a highly localized electric field gradient that is suitable for targeted molecular trapping in solution or inside a cell. b, Scanning electron microscopy (left) and transmission electron microscopy (right) micrographs of the DEP nanotweezer before (top row) and after (bottom row) carbon deposition, d being the electrode gap distance. Scale bars, 20 nm (left column); 100 nm (right column). c, Linear sweep voltammograms recorded for each of the two electrodes (E1 and E2) for a typical nanotweezer using hexaammineruthenium(III) chloride (n = 5 independent measurements). Inset, the distribution of electrode radii calculated from the limiting currents (n = 17 independent measurements). d, Electric field gradient at the nanotweezer tip along the z axis (x = y = 0, fA = 1 MHz, Vpp = 20 V) obtained from finite element modelling (top) and a plot of threshold electric field gradient required to trap double-stranded DNA (bottom). e, Finite element model of the temperature distribution around the nanotweezer tip in different ionic strength solutions.
원문: 여기를 클릭하세요~