(원문: 여기를 클릭하세요~)
과학자들의 지속적 ‘연구 편식’으로 미지의 유전자들 계속 방치, 연구비 지원 방식이 원인으로 분석
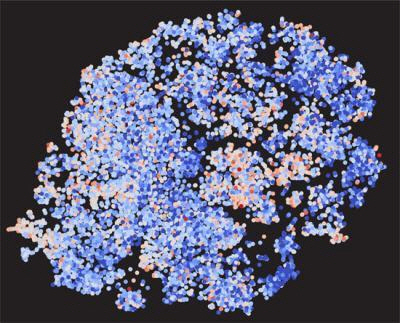
유전자 연구 편향 지도. 점이 유전자이며 파란점은 기대 이하로 연구되는 분야이다./토머스 스퇴거 박사=연합뉴스
15년 전 인간 게놈지도가 이미 완성됐음에도 불구하고, 과학자들의 ‘연구 편식’으로 약 2만개의 인간 유전자 중 90%에 해당하는 1만8,000여개의 유전자가 방치된 사실이 드러났다.
미국 노스웨스턴대학 토머스 스퇴거 박사가 이끄는 연구팀은 2015년까지 발표된 유전자 관련 논문들의 분석 결과, 이런 편향된 연구 경향이 파악됐다고 미국 공공과학 도서관(PLoS) 온라인 학술지 ‘PLoS Biology’를 통해 밝혔다.
연구팀은 인간 유전자 연구가 2,000여 개에만 집중돼 정작 의학적으로 중요한 폐암 관련 유전자나 유방암 유전자군 등에 대한 유전자 연구는 소외되고 있다고 밝혔다. 특히 약 30%에 달하는 유전자들에 대한 연구논문이 단 한 차례도 발표된 적이 없으며, 이런 연구 경향이 지속된다면 100년이 지나도 인간 유전자는 여전히 미지의 영역에 남아있을 것이라고 역설했다. 또한, 이러한 연구 편향은 연구지원 정책이 새로운 유전자 연구 영역을 개척하는 것보다 게놈지도가 완성되기 전인 1980~90년대에 이미 많은 것이 밝혀진 유전자에 대한 추가 연구만을 부추기고 있으며, 갓 박사학위를 딴 연구원들도 기존 토대 연구가 빈약한 유전자 연구에 나서는 모험을 하려 하지 않기 때문인 것이 원인으로 지목됐다.
실제로 이번 연구에서 박사급 연구원과 박사과정 학생이 밝혀진 것이 거의 없는 유전자에 대한 선구자적 연구에 나선 이가, 훗날 독립적 연구자가 될 수 있는 가능성이 그렇지 않은 경우보다 50% 낮은 것이 드러났다. 스퇴거 박사는 이에 대해 “인간 유전자에 관한 현재의 연구들은 의학적 중요도를 반영하지 않고 있다”면서 “인간 질병과 매우 밀접한 관련이 있는 많은 유전자가 아직 연구되지 않고 있으며, 대신 사회적 영향력과 연구비 지원 방식이 현재의 연구를 과거와 같은 주제로 이끌고 있다”고 분석했다. 논문 공동저자인 루이스 아마랄 박사도 “인간 게놈프로젝트로 모든 것이 바뀌었어야 하는데 그대로며, 과학자들은 같은 자리에 똑같은 유전자를 연구 중”이라며 “우리가 모든 관심을 10%밖에 안 되는 유전자에 쏟을 이유는 없다”고 지적했다.
연구팀은 과학자들 스스로 이를 바로 잡을 수는 없으며, 과학계가 안전한 연구에 나서는 과학자보다는 미개척 분야의 유전자를 연구하는 과학자들을 보호하고 육성하는 방향의 노력이 필요하다고 밝혔다.
https://www.theatlantic.com/science/archive/2018/09/the-popularity-contest-of-human-genes/570586/
The Genes That Never Go Out of Style
Since 2003, several researchers have noticed that scientists tend to study genesthat are already well studied, and the genes that become popular aren’t necessarily the most biologically interesting ones. Even among genes, it seems, the rich get richer. This trend hasn’t changed in the past two decades, according to a new study from Thomas Stoeger from Northwestern University. Through a massive analysis of existing biomedical data, he found that he can predict how intensely a given gene is studied based on a small number of basic biochemical traits. Most of these, he says, reflect how easy a gene was to investigate in the 1980s and 1990s, rather than how important it is.
In what Amaral describes as a “heroic effort,” Stoeger spent years collating information from dozens of databases about every known gene. Using machine-learning tools, he then showed that he could accurately predict how many papers have been published about a given gene using just 15 traits.
Some of these telltale traits—how often the gene is mutated, or the negative consequences of losing it entirely—certainly reflect the gene’s importance and its relevance to human disease. They’re the kind of characteristics scientists should be paying attention to.
But other traits—how big the gene is, how active it is, how many tissues it is active in, whether it produces proteins that are secreted from cells, whether those proteins are soluble in water, and more—reflect how amenable the genes are to experiments. Highly active genes, for example, are easier to detect using older methods. “That definitely had a substantial impact on whether you were even able to study a gene in [the 1980s and 1990s],” says Sharon Plon from Baylor College of Medicine. And those historical quirks are better at predicting how the National Institutes of Health currently allocates its money than thousands of other features that more directly reflect what we now know about the role of genes in disease.
It’s possible, of course, that scientists have already identified all the really important genes, and are allocating their attention appropriately. There are good reasons, for example, why p53 is the most popular human gene: It protects our cells from cancer, and is itself mutated in half of all tumors. More broadly, Stoeger found that compared to the least popular genes, the most popular ones are three to five times more likely to have been linked to diseases in large studies, or to wreak havoc when they accrue incapacitating mutations. The problem is that those celebrity genes get over 8000 times more attention than their neglected counterparts. Scientists do tend to study important genes, Stoeger says, but even then, they do so disproportionately.
When will genomics cure cancer?
That’s partly because there are substantial barriers to studying something that no one else has studied before. A researcher might spend years trying to, for example, engineer a line of laboratory rodents that lack the gene in question. They might create bespoke antibodies or other chemical reagents that can help track or visualize the gene. This all takes time, money, and effort. “Many investigators identify an important gene and then spend their whole career studying it,” says Plon.
To do otherwise is risky. Stoeger showed that over the past two decades, junior researchers who focused their attention on the least studied genes were 50 percent less likely to eventually run their own lab. “Those people get pushed out of the biomedical workforce, and then don’t get a chance to set up a lab that explores some of the previously unknown biology,” he says.
Stoeger and Amaral “have done a remarkable job of comprehensively analyzing the reasons why many important genes are ignored,” adds Purvesh Khatri from Stanford University. “Their results underscore the need to change how we study human biology.”
Amaral blames the research imbalance on the erosion of funding from the National Institutes of Health, which forces scientists to compete for a dwindling number of grants and pushes them toward safer research. “When resources stop growing, the entire system is telling people not to take chances,” he says. The NIH does have grants that are meant to promote innovative, exploratory, high-risk research, but even these end up augmenting the same imbalances: Half of the papers that emerge from them still focus on the same 5 percent of well-studied genes. Even supposedly game-changing techniques like crispr have altered the landscape of gene popularity very little. “You get all these new tools but you end up using them on the same set of genes that you were using them on before,” says Amaral.
What if (almost) every gene affects (almost) everything?
Within the past decade, only six genes have escaped the doldrums of obscurity and become newly popular, mainly because researchers recently realized that they are medically important. C9Orf72, for example, was recently identified as a common link between two neurodegenerative diseases—frontotemporal dementia and ALS. IDH1 is commonly mutated in brain cancers. SAMHD1 protects certain cells from HIV. “It’s clear that if sufficiently motivated, the field can tack,” says Shendure, “but I still would have expected more exceptions. We don’t want communism for genes, but we do want to lower the activation energy for intensively attacking the biology of genes that clearly merit more attention.”
Stoeger and Amaral have already created a wish list of genes that, based on their data, should be easier to study with modern methods, and are probably worthy of attention. They also think that agencies like the NIH should create grants that encourage junior scientists to pursue new and unpredictable lines of research, and, crucially, provide them with enough years of funding to offset the initial risk of heading down those paths. “If we don’t take targeted approaches to incentivize the study of unstudied genes, the system is not going to change,” Amaral says.