(원문)
Methods for imaging sugars attached to proteins — the protein glycoforms — are of interest because glycoforms affect protein movement and localization in cells. A versatile approach is now reported that uses DNA as molecular identity tags.
The attachment of sugar molecules to proteins is one of the most common protein modifications, found in all domains of life. Sugars attached to proteins are called glycans, and modulate the physicochemical and physiological properties of the carrier proteins1. But tracking and visualizing glycoforms — the specific patterns of sugars attached to a protein — in cells is challenging, particularly if you want to visualize several different glycoforms at once. Writing in Angewandte Chemie, Li et al.2 now report a method for doing this that relies on the dynamic interactions of a set of DNA codes.
Since the early 1990s, the use of fluorescent tags as labels for proteins has revolutionized how cell biologists analyse protein movement and localization in cells3,4. But even though the types of glycan attached to proteins can affect their movement and localization, it has been difficult to visualize any particular glycoform. One way in which researchers have attempted to solve this problem is by using a technique called fluorescence resonance energy transfer (FRET). In this technique, a fluorescent molecule (a fluorophore) is attached to a protein of interest and a second fluorophore is attached to a specific sugar; fluorescence occurs only if the two molecules come into close proximity through the attachment of the sugar to the protein5–8. However, the need to use two different fluorophores can limit applications, for example by making it difficult to detect multiple glycoforms of a protein in the same experiment.
Li et al. overcome this problem using an approach that they describe as a hierarchical coding strategy, in which multiple single-stranded DNA molecules are used as identification codes to visualize specific sugars attached to a chosen protein (Fig. 1). The first DNA molecule used in the authors’ system contains a sequence (known as an aptamer) that specifically binds to the target protein. The aptamer is attached to another sequence (the protein code) that identifies the protein. A second DNA molecule, called the timing code, contains a sequence that is complementary to the protein code, and that therefore hybridizes (forms a double helix) with it.
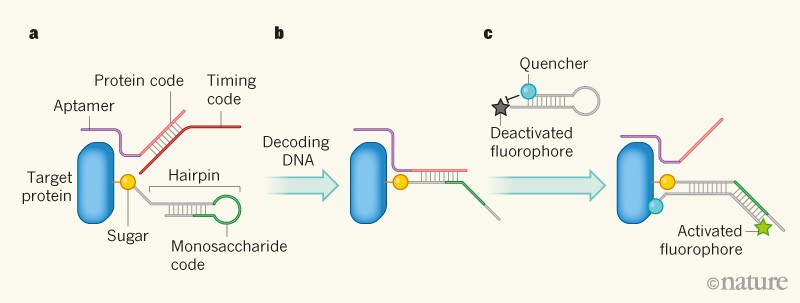
Figure 1 | A method for visualizing sugars on proteins2. a, A DNA sequence (the protein code) is bound to the target protein through a sequence called an aptamer. The protein code is hybridized (forms a double helix) with a second sequence, called the timing code. A sugar attached to the protein is covalently attached to a ‘hairpin’ DNA, which contains a masked sequence that identifies the sugar (the monosaccharide code). b, A ‘decoding’ DNA molecule is added that hybridizes with the timing code (not shown), releasing the protein code so that it hybridizes with part of the hairpin. The hairpin opens, unmasking the monosaccharide code. c, A second hairpin DNA is added, which is complementary to the monosaccharide code; it also bears a fluorescent molecule (a fluorophore) at one end and a quencher molecule at the other, which deactivates fluorescence. The hairpin hybridizes with the DNA containing the monosaccharide code, opening the hairpin and allowing the fluorophore to fluoresce. The protein code is simultaneously exposed by this process, and can take part in another cycle of reactions.
The third DNA molecule used in Li and colleagues’ system contains three segments. The first segment is complementary to the protein code. This is attached to a second sequence called the monosaccharide code, which identifies a specific sugar. The third segment has a sequence that enables the complete strand to form a structure known as a hairpin, which masks the monosaccharide code. The hairpin DNA is covalently attached to the sugar identified by the monosaccharide code. If the hairpin-bearing sugar is in turn attached to the target protein, this can bring the hairpin into close proximity with the double helix formed by the protein and timing codes.
The final key component of Li and colleagues’ system is another hairpin DNA, which contains a complementary sequence to the monosaccharide code and a sequence that can displace the protein code from a double helix. The hairpin also has a fluorophore attached at the 5ʹ end, and a ‘quencher’ molecule at the 3ʹ end. The quencher stops the fluorophore from fluorescing when the hairpin is closed, but allows fluorescence when the hairpin opens.
So how do all these components interact to decode the crucial DNA identifiers and allow glycoforms to be visualized? The process is triggered when a single-stranded DNA that is complementary to the timing code is added to the system. This DNA hybridizes with the timing code, thus displacing and exposing the protein code. The exposed protein code then hybridizes with the complementary sequence in the hairpin attached to the sugar, opening up the hairpin and unmasking the monosaccharide code.
When the fluorophore-carrying hairpin is added to the system, the unmasked monosaccharide code hybridizes with the complementary DNA sequence in that hairpin. The hairpin therefore opens up, allowing its fluorophore to fluoresce: in effect, a fluorescent tag has been attached to the sugar, allowing it to be detected. The hybridization also unmasks the protein code, making it available for another reaction cycle. The key element of Li and colleagues’ system is that the protein code is physically associated with the target protein, because this ensures that only hairpin-bearing sugars that are attached or close to the protein can become fluorescent.
The authors confirmed that the chain of reactions occurs in a cell-free system in vitro and used it to identify two glycoforms of the MUC1 protein: MUC1 decorated with the sugar fucose, or with another sugar called sialic acid. Crucially, the authors also showed that the fluorescent signals can be generated and detected on cells that had been modified using a method known as metabolic labelling9 to incorporate hairpin-bearing sugars.
An advantage of this method is that, because the choice of DNA sequences that can be used as labels is effectively infinite, many different glycoforms can be imaged, as long as the proteins and sugars can be specifically labelled with their own DNA codes. Moreover, the authors clearly showed that sialylated and fucosylated MUC1 could be simultaneously detected using their method. One potential limitation, however, is that the DNA used was not observed to be transported into cells through natural processes, suggesting that intracellular glycoforms cannot be detected by this method. This could actually be an advantage for studies that focus on cell-surface proteins.
A few issues will need to be clarified in future studies. For example, the efficiency of the decoding process is unclear. It is also not known whether sugars on molecules next to the target proteins might sometimes become fluorescent, as a result of DNA hybridization between the protein code and hairpins attached to sugars on neighbouring proteins. Because a large number of glycans are attached to MUC1, the method might not need to be highly efficient to generate a detectable fluorescent signal for this protein, and any minute signals produced from neighbouring molecules would not be a serious problem. However, further experiments using other glycoproteins that have fewer sugars attached are needed to validate the method fully.
Given that both the above issues might depend largely on the length of the DNA chains used, careful design of the DNA codes and of the aptamers will be essential for ensuring the specific detection of other glycoforms. The practical advantages and disadvantages of the new technique compared with other strategies for glycoform imaging that have been reported in the past few years — including two methods reported by workers from the same group as Li et al.10,11 — also remain to be explored.
Nevertheless, Li and colleagues’ hierarchical coding strategy for glycoform imaging shows great potential, and could be an important step in the development of a system analogous to the use of green fluorescent proteins for protein tagging — which is now standard practice for biologists. The ultimate goal is to visualize glycoforms in a way that will enable us to see what we want to see, rather than only what can be seen.
Nature 561, 38-40 (2018)