(원문)
Genome-editing approaches have been used to fuse 16 yeast chromosomes to produce yeast strains with only 1 or 2 chromosomes. Surprisingly, this fusion has little effect on cell fitness.
The genomes of nucleus-bearing organisms are divided into linear chromosomes. The number of chromosomes ranges from one to hundreds across species. But why is there such variation? Do specific chromosome numbers hold an advantage for particular species? In two papers in Nature, Shao et al.1 and Luo et al.2 independently manipulate the genome of the budding yeast Saccharomyces cerevisiae by systematically fusing chromosomes, enabling the researchers to explore the consequences of chromosome-number reduction.
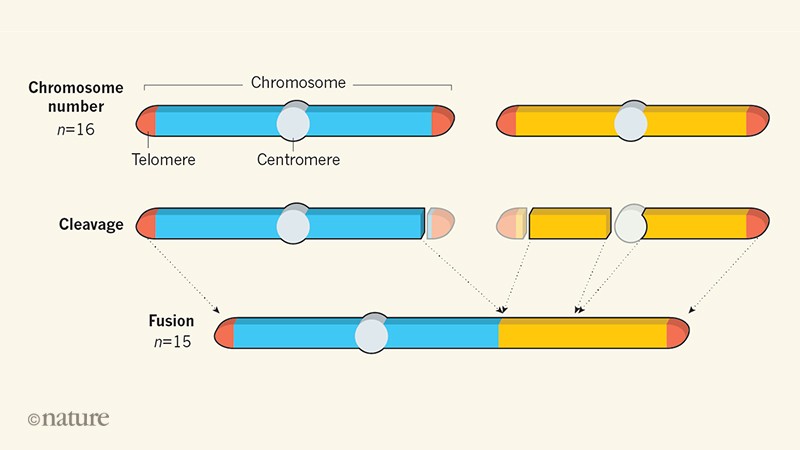
Normal S. cerevisiae genomes have 16 distinct chromosomes (n = 16), which range from 230 to 1,532 kilobases in length3. To function correctly, yeast chromosomes need protective structures called telomeres at both ends, and only one centromere — a region that ensures the accurate segregation of chromosomes into mother and daughter cells during cell division. Simply fusing the ends of two chromosomes is therefore not a viable strategy for reducing chromosome number because it would produce chromosomes containing two centromeres.
To solve this problem, the two groups used genome-editing tools to fuse sequences found adjacent to one of the telomeres in each chromosome, and to simultaneously remove one of the two centromeres (Fig. 1). Using this approach, they reduced the chromosome number step by step, producing strains that had progressively lower values of n. The fusion strains comprised genomic material that is almost identical to that of normal S. cerevisiae, differing only in chromosome number and by a few non-essential genes that were deleted during strain creation.
Figure 1 | Fusing chromosomes one by one. Two groups1,2 have fused all 16 chromosomes (n = 16) of the budding yeast Saccharomyces cerevisiae to produce strains that have only one1 or two2 long chromosomes. In S. cerevisiae, each chromosome must have protective structures known as telomeres at both ends, as well as a single structure called a centromere that is essential for normal chromosome segregation during cell division. To generate fused chromosomes with this composition, the groups used genome-editing techniques to cleave sequences found next to one of the telomeres in each chromosome, and simultaneously removed one of the two centromeres (just one cleavage site is sufficient to trigger removal of the entire centromere). They then fused the cleaved portions. By repeating this process, they reduced the chromosome number in a stepwise manner, producing yeast cells that have progressively lower values of n.
Luo et al. produced an n = 2 strain containing chromosomes that were each about 6,000 kb long. However, they were unable to fuse the two chromosomes into one as part of a viable cell. By contrast, Shao et al.successfully fused the entire S. cerevisiae genome into a single chromosome in a functional yeast.
Given that each group used similar strategies, it is interesting to consider why only one of the teams could fuse the final two chromosomes. A possible explanation is that the groups fused the yeast chromosomes in different orders and orientations. Perhaps such factors matter, which could mean that only certain final genome structures are attainable. In the future, reducing the chromosome number through a variety of fusion paths might reveal how chromosomal structures affect cell viability. Another possibility is that mutations introduced accidentally during the chromosome-fusion experiments affected cell tolerance to the new genome organization.
Both groups then investigated the biological implications of chromosome fusion. Overall, organismal traits such as cell growth, size and shape seem to be buffered throughout the series of fusions. Notably, the expression of only a few genes was altered considerably in either the n = 2 or n = 1 strains. Most of the observed increases in gene expression can be explained by there being fewer genes located near telomeres, which promote transcriptional silencing4.
Such transcriptional stability is in contrast to the widespread transcriptional variation that is seen when yeast undergoes other types of chromosomal modification, such as inversions of particular regions5. Shao et al. show that this stability reflects the fact that there are only modest changes to the intrachromosomal interactions that usually take place, which can modulate gene expression. However, the interchromosomal-interaction landscape changes drastically, owing to the depletion of centromeres, which drive the 3D configuration of the yeast genome6.
The yeast strains generated by the groups are haploid — they contain only one copy of each chromosome. Haploid yeast reproduce asexually, but they can also mate through sexual reproduction to form diploid yeast, which contain two copies of each chromosome. Diploid yeast can then divide through a process called meiosis to produce haploid spores that mature into haploid cells. The groups showed that the n = 1 and n = 2 strains can undergo sexual reproduction, albeit with reduced efficiency compared with wild-type yeast, and produce spores that are slightly less viable.
During meiosis, genetic material is exchanged between paired chromosomes in a process called recombination. Because the genomes of all cells from a given fusion strain are identical, they lack the genetic variability that researchers need to map recombination through the generations. As such, the two groups could not characterize how chromosome reduction affects recombination. The high spore viability of each fusion strain indicates that some recombination might occur, ensuring proper chromosome segregation. However, the greatly reduced chromosome number essentially eliminates any risk of mis-segregation.
Luo et al. mated strains that had different chromosome numbers, and then investigated spore viability and production in the resulting hybrid strains, to determine at what point the fusion strain could no longer produce viable spores (a phenomenon called reproductive isolation). As predicted7, an increasing difference in chromosome number had an increasing effect on spore viability until, in hybrids generated by crossing haploid strains that have n = 16 and n = 8, none of the spores produced were viable. Moreover, spore production was arrested when the difference in chromosome number became any larger.
This is unexpected, especially given that diploid hybrids that are sterile because of high sequence divergence or differently arranged genomes between their two sets of chromosomes can progress efficiently through meiosis, despite producing inviable spores7. The mechanism that underlies the reproductive isolation seen by Luo and colleagues remains to be determined. Future work using synthetic genomes, which can be edited at the single-nucleotide level, will allow the introduction of genetic variants on both local and genome-wide scales, enabling the in-depth, systematic analysis of the factors that prevent species from breeding, as well as the genomic changes that prompt reproductive isolation.
Both studies concluded that reduced chromosome number causes no major growth defects when cells are grown under various conditions and stresses. Small fitness defects were most evident in the n = 1 strain, consistent with the fact that this chromosome configuration is challenging to obtain. Although these fitness differences seem mild in a laboratory setting, they could become more harmful in the natural environment. Indeed, Shao and colleagues’ n = 1 strain was quickly outcompeted by a normal strain of S. cerevisiae when the two were cultured together. This is consistent with the idea that the structure of S. cerevisiae chromosomes has remained highly stable for several million years8,9, although reductions in chromosome number through telomere fusion and centromere loss occurred repeatedly over longer evolutionary timescales10.
The short generation time of S. cerevisiae means that, in the future, the evolution of strains that have a reduced chromosome number could be tracked in the lab, in long-term experiments that run for months or years. Such experiments will enable researchers to map adaptive changes that restore fitness in strains that have a reduced number of chromosomes, and to accurately measure genome stability in these yeast.
Beyond the current findings, these engineered yeast strains constitute powerful resources for studying fundamental concepts in chromosome biology, including replication, recombination and segregation. The chromosome-engineering approach might also be applicable to organisms that have more-complex genomes. However, the presence of highly complex DNA sequences in the regions that surround telomeres and centromeres in these organisms will make this a challenging task.