(원문)
Nature Chemistry volume 10, pages 802–803 (2018)
A new pyrrolysyl-tRNA synthetase/PyltRNA (PylRS/PyltRNA) pair that is mutually orthogonal to existing PylRS/PyltRNA pairs has now been discovered and optimized. This system could enable the site-specific incorporation of a greater number of distinct non-conical amino acids into a protein.
The cellular machinery responsible for synthesizing proteins has evolved to incorporate the correct amino acid into a protein’s polypeptide chain with extremely high fidelity. This poses a problem for anyone wishing to harness the process to incorporate non-conical amino acids (ncAAs) to create designer proteins. One common route to achieve this is using a modified pyrrolysyl-tRNA synthetase (PylRS) and tRNA pair. PylRS is an enzyme responsible for charging tRNA with pyrrolysine encoded by the ‘amber’ UAG stop codon. The PylRS/PyltRNA pairs originated from Methanosarcina mazei (Mm) and Methanosarcina barkeri (Mb) are orthogonal to endogenous aaRS/tRNA pairs in both prokaryotic and eukaryotic cells, which enables them used in conjunction with aaRS/tRNA pairs without affecting the incorporation of canonical amino acids into a protein.
Since their discovery, the PylRS from Mm and Mb have been systematically evolved by researchers and have become the most widely used system to genetically encode a vast variety of non-canonical amino acids (ncAAs) into a diverse range of proteins, including within living organisms1,2,3,4. The technique has led to a burst of new methods involved in probing, imaging, profiling as well as manipulation of proteins within their native biological context. However, whereas incorporation of one ncAA into a protein of interest is efficient and relatively simple, an upgrade of the system to simultaneously incorporate multiple ncAAs still remains challenging.
A single protein chain carrying multiple ncAAs would facilitate more intricate applications such as Förster resonance energy transfer studies5, but requires the presence of additional aaRS/tRNA pair that is mutually orthogonal to current ones. In previous work, Jason Chin and co-workers pioneered the efficient incorporation of multiple ncAAs into proteins. To do this they developed an orthogonal translation system that contains an engineered ribosome named Ribo-Q that can decipher both amber codons and quadruplet codons6. In addition, the PyltRNA was mutated to recognize the ‘ochre’ UAA codon or ‘opal’ UGA codon7. These developments allowed the utilization of the tyrosine synthetase/tRNA pair found in Methanococcus jannaschii(MjTyrRS/MjTyrtRNA) as an orthogonal partner with the MmPylRS/MmPyltRNA pair to incorporate two distinct ncAAs, but this requires the simultaneous suppression of two out of the three ‘stop codons’ within a cell, which can be highly toxic.
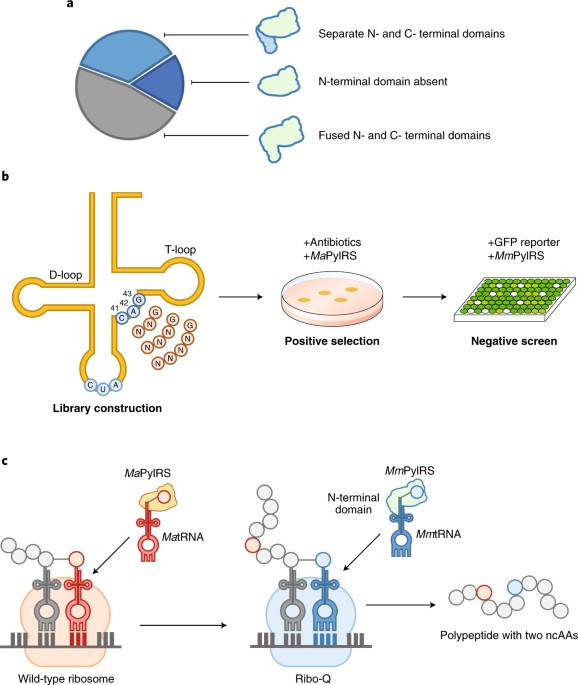
Finding a new PylRS/PyltRNA pair with comparable ncAA incorporation efficiency and complete orthogonality to their homologues in Mm and Mb is challenging, nevertheless, writing in Nature Chemistry, Jason Chin and colleagues have now reported such a pair9. The N-terminal domains of MmPylRS and MbPylRS bind to the T-arm and variable loop of PyltRNA, and are essential to the enzyme activity8 (Fig. 1a). Since tRNAs of pyrrolysine have similar sequence and structures, it is expected that MmPylRS can extensively bind to PyltRNAs of another species. Another class of PylRS have their N-terminal domains encoded separately in different genes. Based on these findings, Chin and co-workers focused their search on this third class of PylRS enzymes that do not possess a distinct N-terminal domain as shown by Mm and Mb. First-round candidates were selected by comparing the sequence similarities to MmPlyRS. After that, genomes that contain a distinct PylRS N-terminal domain-like sequence were removed from the pool. At the end, five PylRS sequences were identified along with their cognate tRNAs in the genomes9. Among them, PylRS from Methanomethylophilus alvus (Ma) has the highest ncAA incorporation efficiency, though MaPyltRNA can be recognized by MmPylRS. By contrast, PylRS from Methanogenic archaeon ISO4-G1 (G1) is mutually orthogonal to the MmPylRS/MmPyltRNA pair.
a, Classification of PylRSs based on (i) the presence of a N-terminal domain, (ii) the presence of a separate N-terminal domain-like protein and (iii) the absence of a N-terminal domain. b, Evolution of MaPyltRNA. tRNA libraries were created by randomizing and extending the variable loop. A positive selection was performed in the presence of MaPylRS, followed by a negative screen in the presence of MmPylRS. c, Two distinct ncAAs were encoded using the mutually orthogonal PylRS/PyltRNA pairs. The MaPylRS/MaPyltRNA pair was assigned to the amber codon, whereas the MmPylRS/MmPyltRNA pair was assigned to a quadruplet codon. Panels a and b adapted from ref. 9, Macmillan Publishers Ltd.
Instead of finding ways to enhance enzymatic activity of G1PylRS, the team decided to focus on improving the orthogonality of MaPylRS/MaPyltRNA pair. Lacking the N-terminal domain responsible for binding the variable loop of MaPyltRNA, this variable loop of MaPyltRNA can indeed be reprogrammed. A tRNA library was created by replacing the nucleotides at positions 41, 42 and 43 on MaPyltRNA with random ones. A positive selection was performed to pick up mutants that can be recognized by MaPylRS, followed by a negative screen to eliminate sequences that are recognized by MmPylRS or other endogenous aaRSs (Fig. 1b). Looking at all of the positive results, the team noticed that the only observed pattern is a conserve G at position 43. Next, a bigger tRNA library was then created with the variable loop lengthened to four, five and six nucleotides, holding G43 constant. The champion went to the tRNA bearing an ‘AUAG’ sequence at the variable loop. The new MaPyltRNA is mutually orthogonal to the MmPylRS/MmPyltRNA pair, with up to 91% of enzyme activity retained.
MmPylRS and MbPylRS have previously been evolved by mutagenesis at their catalytic domains to accommodate ncAAs with different chemical structures. For instance, mutations on MmPylRS can be transplanted to MbPylRS for the incorporation of the same ncAA. Through aligning the binding pocket sequence of MaPylRS to that of MmPylRS, corresponding mutations could be identified on MaPylRS. The resulted enzyme selectively incorporates the specific ncAA but not any natural amino acids or other ncAAs. To demonstrate this the team encoded two distinct ncAAs on one single polypeptide. A MaPylRS/MaPyltRNA pair and the quadruplet anticodon version of MmPylRS/MmPyltRNA pair were introduced into cells that produce Ribo-Q, along with a protein encoded by a gene that contains an amber stop codon and a quadruplet codon (Fig. 1c). Addition of both ncAAs corresponding to each PylRS was required to express the protein in full length.
The discovery of MaPylRS/PyltRNA has expanded the inventory of mutually orthogonal synthetase/tRNA pairs and integration of these mutually orthogonal aaRS/tRNA pairs may allow the incorporation of a greater number of different ncAAs into a single protein in vivo — and thus the installation of a greater range of new and unnatural functionalities. Such a systematic endeavour may ultimately lead to the biosynthesis of fully unnatural polypeptides in Escherichia coli. Huge progress has been made in computational protein design and in the future, de novo design of proteins seems highly probable. The aid of orthogonal PylRS/PyltRNA pairs will enable a wider range of amino acids to be embedded into newly designed proteins. In addition, the orthogonality of these synthetase/tRNA pairs and their performances in eukaryotic cells and multicellular organisms is another interesting question that will need to be systematically explored. Furthermore, the complexity of the system and operational difficulty escalates with the number of ncAAs incorporated. More efforts must be made to lower the prerequisite specialist techniques and skills to enable these approaches to be deployed as a common platform available to most laboratories. Finally, the discovery of mutually orthogonal PylRS/tRNA pairs for the same amino acid may raise great interests in exploring and studying the phenomenon of mutual orthogonality across different species. It gives a perspective on how molecular machineries from mutually orthogonal organisms can be harnessed as powerful tools to disentangle complicated biological processes.