(원문)
Nature Chemistry volume 10, pages 800–801 (2018)
Enzymes can perform various biological functions because of their delicately and precisely organized structures. Now, simple inorganic nanoparticles with a rationally designed recognition capability can mimic restriction enzymes and selectively cut specific DNA sequences.
Designing nanomaterials with properties that enable them to mimic the components of biological processes is a challenging task. Inspired by the passion of scientists to control biological mechanisms at the nanoscale, the development of nanomaterials that mimic enzyme composition and functionality is indisputably one of the frontiers of modern nanoscience1,2,3,4,5. For example, inorganic nanomaterials such as iron oxide, cerium oxide and copper hydroxide nanoparticles have been proposed for use as artificial enzymes, bridging the gap between biological and inorganic materials6,7,8,9. One particularly exciting avenue is the design of artificial nanomaterials that mimic restriction enzymes. Restriction enzymes are an irreplaceable tool in genetics, molecular biology and therapeutics, which recognize and cleave specific DNA sequences at predetermined sites. A new class of nanomaterials that possess such capabilities would facilitate the sequence-specific cutting of nucleic acids and can potentially be used in targeted therapies.
A major advance in this direction has now been reported in Nature Chemistry10 in a study led by Chuanlai Xu and Hua Kuang at Jiangnan University, China. The study reports on an artificial biomimetic DNA endonuclease that can cut the phosphodiester bonds — which link the sugar moieties in the backbone of nucleic acid strands, much like its biological counterpart.
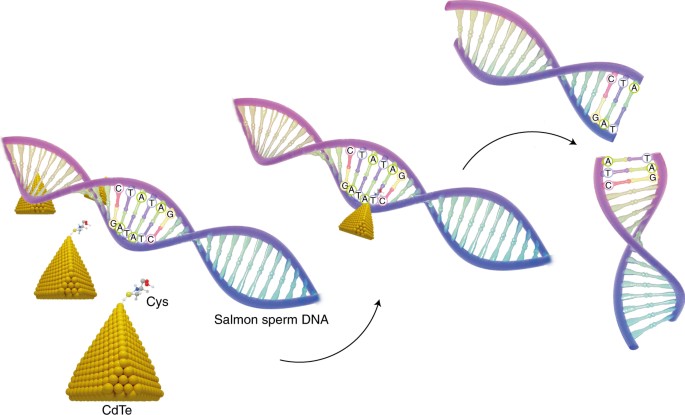
The team synthesized semiconducting cadmium telluride (CdTe) nanoparticles with a truncated tetrahedral shape, which were coated with chiral cysteine. These coated nanoparticles selectively bind to DNA of a certain length (above 90 base pairs) and can recognize the DNA sequence GATATC. Upon excitation with polarized light, a nanoparticle attached to the DNA sequence acts as a photosensitizer and produces the hydroxyl radical that cuts between the GAT and ATC units (Fig. 1). The experiments revealed that the nanoparticles acted as an electron donor inducing the generation of reactive oxygen species, while DNA acted as the receptor, which led to cleavage of the phosphodiester bond. Usually, classical methods to perform DNA scission are based on the hydrolytic cleavage through the hydrolysis of phosphodiester linkages and the oxidative scission of the deoxyribose residues through a redox reaction.
Semiconducting CdTe nanoparticles with a truncated tetrahedral shape, were coated with chiral cysteine (Cys). These inorganic nanomaterials specifically recognize the GATATC restriction site in a DNA sequence. When illuminated with circularly polarized light the CdTe nanoparticles act as a photosensitizer and cut the DNA sequence between the GAT and ATC units, mimicking a restriction endonuclease.
Modelling conducted within this study also confirmed preferential adsorption of bases A and T on the same groove along the edge of the tetrahedral nanoparticles, whereas C and G were found preferably in the nearest corner. An array of experiments with control sequences and nanoparticle functionalization chemistries revealed the specificity of the approach, both in terms of the composition and the location of the sequence. Chiral-cysteine-modified nanoparticles could efficiently cut DNA, whereas nanoparticles modified with other ligands had no obvious effect. Importantly the activity of the artificially created endonuclease was explored in complex physiological environments, where photoactivated DNA cutting was shown to occur both in living cells and in vivo.
Although novel strategies using nanoparticle as carriers for gene-editing tools such as CRISPR have been recently reported11, the innovation in this study is in the design of nanomaterials that carry out the cutting themselves. The latter effect could be exceptionally beneficial in therapeutic applications when you take into account the large number of existing strategies to deliver nanoparticles in vivo.
The work demonstrates that chiral materials represent a new pathway for constructing exquisitely designed inorganic enzymes made from nanoparticle building blocks. Creating and understanding the mechanisms of cysteine-functionalized CdTe nanoparticles can potentially enable the design and development of a further class of artificial enzymes to better control and understand DNA cutting in biological systems. Although the manuscript only reports on a single type of nanoparticle capable of recognizing and cutting the GATATC DNA sequence, the present study will probably shed light on interactions between DNA and chiral nanomaterials and on future nanoparticle designs that can target alternative DNA sequences. This opens up the possibility of synthesising a cocktail of inorganic enzymes which can provide a more complete toolbox for gene-editing applications. Such a development would have substantial benefits for the development of new tools in gene analysis and therapies, and would have a significant impact in basic research and potentially in therapeutics.