Synthetic genetic circuits leverage the information processing capability of biological machinery to tackle complex sensing tasks, yet they lack many of the advantages inherent to electrical computation. Now, an interface has been designed that provides an electrical output for synthetic genetic circuits.
Synthetic genetic circuits are engineered collectives of nucleic acids and proteins that act cooperatively in response to biorecognition events, by triggering downstream processes and reaction cascades, to accomplish bioproduction, logical operations, memory storage and sensing1. Gene circuits can perform several of the functions of their electronic counterparts — response to multiple inputs, signal amplification and decision-making — and can be housed within living cells2. However, cell-free versions carry the advantage of more robust storage and fewer biosafety regulations3. Logic is encoded by the biological equivalent of ‘if, then’ statements and Boolean arguments. The prevailing method to extract information from gene circuits is transduction of the output signal to give a change in fluorescence via expression of a fluorescent protein, enzymatic activation of a small-molecule fluorophore or turn-on of Förster resonance energy transfer probes.
Current strategies for reporting the output of a sensing circuit have two important limitations. First, if the only communication between the gene circuit and external instrumentation occurs at the final output, then all of the logic that occurs between sensing and reporting must be molecularly encoded, which is a formidable challenge. Second, until now, the depth of information that is able to be extracted is limited by the different number of colours available for fluorescence (typically, five). Now, writing in Nature Chemistry, a team led by Shana Kelley and Keith Pardee report that they have addressed these limitations by developing an electrochemical interface that connects cell-free, gene-circuit-based sensors and provides an electrical output4.
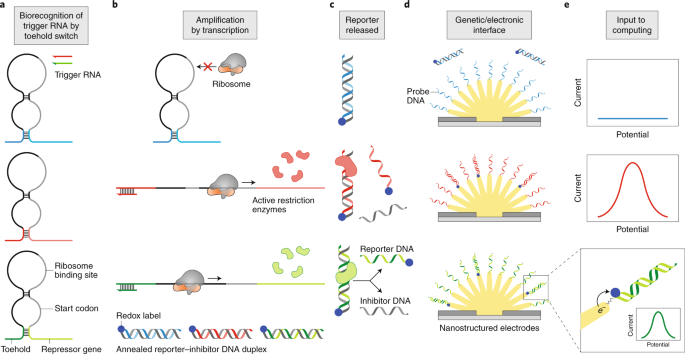
Figure 1 illustrates the concepts underlying the operation of the gene circuit/electrode interface. The process is initiated when a biorecognition event activates a biomolecular reaction. In this case, first, the specific binding of a target RNA strand to its complement, a hairpin-looped RNA, opens the loop to make it accessible. This recognition motif is called a toehold switch (Fig. 1a) and is a gene regulator designed for synthetic biology5. Second, a ribosomal complex binds to the exposed strand and transcribes it to produce a restriction enzyme (Fig. 1b). Importantly, many enzymes are produced, thereby yielding an amplified response. Third, the restriction enzyme cleaves double-stranded DNA in a sequence-specific manner (Fig. 1c). This DNA duplex is key to the strategy developed by the Kelley and Pardee group because one of the DNA strands (the reporter) is tagged with a redox-active molecule that can be detected electrochemically. Each cleavage event liberates one such single-stranded reporter DNA from a complementary inhibitor DNA strand, and in this way, redox-labelled oligomers are accumulated for subsequent detection. Fourth, each labelled strand binds to a nucleic acid probe on an electrode surface, thereby bringing the redox molecule into sufficiently close proximity to the electrode for detection (Fig. 1d). This electrode is nanostructured to increase its surface area, thereby enhancing sensitivity. Finally, an electrical potential, applied to the electrode by a potentiostat, drives an electron transfer reaction at the electrode, leading to an electrical current that is proportional to the surface coverage of the redox molecule (Fig. 1e). The result is an amplified and quantitative electronic readout of the gene circuit.
a, The gene-circuit sensor takes an analyte as its input. If the trigger RNA binds to the toehold, then the hairpin loop opens. b, Opening the hairpin loop makes a repressed gene accessible for transcription, leading to the production of a restriction enzyme. Each activated toehold switch results in the generation of many enzymes, and in this way, the signal is amplified. c, The restriction enzymes cleave annealed reporter–inhibitor DNA duplexes in a sequence-specific manner and, by this process, liberate reporter DNA. d, Binding of this reporter DNA to a surface-bound probe DNA brings the redox label into sufficiently close proximity to the electrode to allow electron transfer. e, The current resulting from this electrochemical reaction is input to an electronic circuit for computing.
Their approach retains the advantages of a gene circuit while providing a convenient ‘off ramp’ to an electronic circuit, thereby introducing design flexibility with regard to the division of computing tasks. Furthermore, the number of possible outputs is increased because the information is spatially encoded in an array of electrodes. This enables each distinct probe to be connected to a separate electrode and so the output of different probes can be easily monitored. This also breaks the requirement to have a different (observable) fluorophore for each output channel. The robust cell-free format and low-cost electrochemical equipment are especially well suited to point-of-need applications. These advancements have the potential to dramatically increase implementation of gene-circuit-based sensors to address pressing needs in human health.
A design feature that increases the relevance of this approach is that the biomolecular/electronic interface, which comprises the restriction enzymes, reporter–inhibitor duplex DNA and surface-modified electrodes, is universal as the probe sequence can be customized to bind any reporter. Meanwhile, the upstream biorecognition and amplification schemes as well as the downstream computer processing can be readily adapted to many specific sensing applications. The team demonstrated the versatility of their approach by altering the recognition step. In this experiment, the expression of a restriction enzyme was inhibited by the protein tetracycline repressor, which is subsequently released when bound by the small molecule anhydrotetracycline.
Multiplexed sensing was accomplished by employing parallel gene circuits, each comprising a distinct set of toehold switch, restriction enzyme, reporter–inhibitor duplex DNA and probe-modified electrode. The team describe the process by which they systematically selected each component of the gene circuits to maximize speed and sensitivity while avoiding crosstalk. First, they screened 66 commercial restriction enzymes, selecting the 10 enzymes with the highest rate of expression, highest DNA cleavage activity and lowest cross-reactivity. These features were evaluated using a Förster resonance energy transfer probe for which fluorescence turned on when the probe was cut. Next, successful generation of electrochemical signal at the matched electrodes for individual and mixed enzymes was demonstrated. Finally, cross-reactivity of the restriction enzymes with the DNA encoding them was also evaluated and mitigated by methylation or editing of the DNA sequence where needed. The performance of six of these toehold-switch gene circuits was then showcased by detection of six synthetic RNA strands, which resulted in sequence-specific electrochemical signals.
Kelley, Pardee and co-workers demonstrated the potential impact of these multiplexed gene-circuit-based electrochemical sensors in a timely application — the detection of four resistance genes for the last-line antibiotic colistin. First, they developed and optimized toehold-switch sensors paired with four of the top-performing restriction enzymes. Next, the team demonstrated both individual and multiplexed detection of the genes. Finally, these gene circuits were paired with the isothermal amplification technique nucleic acid sequence-based amplification to achieve a low femtomolar detection limit for a resistance gene.
Antibiotic resistance has been identified in livestock and is a global issue that requires rapid and portable methods of detection. The approach designed by Kelley, Pardee and co-workers is compelling because this gene circuit/electrode interface is particularly well suited to point-of-need applications, allowing frequent evaluation and immediate intervention. The components of the cell-free gene circuit can be freeze dried for long-term storage and transport, and further, electrochemical instrumentation is less expensive and more compact than even the smallest fluorescence detectors. Finally, the team’s choice of isothermal amplification boosts sensitivity without incurring the added complexity of thermal cycling.
This important work lays the foundation for several future advances. First, to capitalize on the inherent advantages of the design for ‘sensing at the point of need’, the platform needs to be re-designed to make use of inexpensive and durable materials that are readily manufactured. Such a transformation would expedite translation to practical applications such as point-of-need diagnostics. Second, the signals reported here are in the range of tens of nanoamperes, which is a relatively challenging measurement for a point-of-need device. Furthermore, nucleic acid amplification is required to obtain such a signal for relevant quantities of resistance genes. Therefore, further strategies or development are needed to improve sensitivity. While the electrochemical strategy employed here — binding of a redox-labelled DNA reporter to an electrode-bound probe — is effective, several more sensitive schemes involving multiple redox labels or catalytic amplification have been reported6 (some by Kelley’s own group) and could be leveraged in this context. Finally, due to the universal nature of the biological/electronic interface reported here, there are many opportunities to tailor upstream (gene circuit) and downstream (electronic circuit) functions to accomplish specific sensing goals. For instance, computation using electrical circuits could be used to design more complicated logic networks without requiring the design of an interface between two (or more) molecular reaction networks that do not share any molecular components. The development of more complicated molecular circuits, such as the introduction of aptamers, could broaden the range of analytes that can be detected and therefore open up more applications, for example, multiplexed detection of whole pathogens and small-molecule drugs. This work is, therefore, expected to have a far-reaching impact.
(원문: 여기를 클릭하세요~)