Abstract
Monogenic disorders occur at a high frequency in human populations and are commonly inherited through the germline. Unfortunately, once the mutation has been transmitted to a child, only limited treatment options are available in most cases. However, means of correcting disease-causing nuclear and mitochondrial DNA mutations in gametes or preimplantation embryos have now been developed and are commonly referred to as germline gene therapy (GGT). We will discuss these novel strategies and provide a path forward for safe, high-efficiency GGT that may provide a promising new paradigm for preventing the passage of deleterious genes from parent to child.
Main
There are over 10,000 identified single-gene (monogenic) mutations so far that are linked to human diseases. A genetic mutation is typically presented as an alteration of the genetic code (or sequence) that ultimately results in abnormal proteins or other gene products. While some gene mutations are rare, the cumulative effect of thousands of different gene mutations on human populations is immense, affecting hundreds of millions of people worldwide. Genetic conditions are also a leading cause of early pregnancy loss, infant mortality and birth defects1.
Many gene mutations are passed down to offspring (so-called heritable, or germline, mutations) and thus persist in certain human lineages and cause familial diseases (Box 1). The high frequency of genetic disorders has spurred an interest in novel strategies for prevention of the transmission of pathogenic mutations from parent to child. While germline gene therapy (GGT) has proven to be an ethically sensitive subject, exploration of therapeutic possibilities of GGT is not premature given the dramatic evolution of the molecular diagnostics of most genetic disorders, and the ability to correct these defects in the germline2.
Experimental gene therapy approaches that involve treatment of affected somatic tissues or organs in patients are referred to as somatic gene therapy. However, most pathogenic gene mutations, once passed down to children, incite irreversible damage to tissues and organs and cause diseases such as hypertrophic cardiomyopathy, Huntington’s disease or BRCA-associated cancers. In these cases, somatic gene therapy is unlikely to reverse the pathology of the disease unless therapy is attempted earlier, before onset of disease. There have been proof-of-concept studies in mouse models of genetic disease that have demonstrated that the success of fetal gene therapy for selected congenital genetic disorders, such as tyrosinemia type 1 and neuronopathic Gaucher disease, is possible3,4. The efficacy of somatic gene therapy, if feasible at all, is also complicated by the necessity of targeting billions of cells in solid tissues and organs. Lastly, confining treatments to somatic cells of individual patients will not prevent transmission of defective genes from parents to children.
Gene therapy directed at reproductive cells (sperm and eggs) or preimplantation embryos, termed GGT, has the potential to correct disease-causing mutations to wild-type variants early in development when the mutation is present in one of few embryonic cells. In addition, GGT will not only prevent passage of genetic disease to a child, but to all future generations.
Gene therapy in general utilizes several alternative strategies that were initially developed for clinical applications on somatic tissues. One of the first approaches involves inserting a synthetic copy of a normal gene into a nonspecific location within the genome in addition to the mutant, endogenous copy (or copies) of the gene. The transgene is usually delivered and integrated into the target genome using viral vectors. This approach is undesirable for GGT due to safety and efficacy concerns related to using viruses and random integration of transgenes. In addition, this form of gene therapy results in genetic modifications of the human genome that are deemed ethically unacceptable for GGT.
Another alternative approach is based on inducing lesions in genes that are already mutated using genome editing in hopes of altering the gene to modulate disease. Genome-editing tools, including RNA-guided CRISPR–Cas9, are highly effective for site-specific DNA breaks leading to targeted insertion and deletion (indel) mutations at specific loci in animals and human cells (see details below)5. There are a few cases in which the generation of novel mutations has been proposed for therapeutic applications in somatic cells. In particular, strategies to modify mutations causing Duchenne muscular dystrophy (DMD) were tested in mouse models. Cas9-mediated excision of exon 23 of the dystrophin (Dmd) gene, which harbors the nonsense mutation responsible for the mouse DMD phenotype, restored expression of a truncated version of the dystrophin protein and improved skeletal muscle strength to varying degrees6.
Another example is that of the C–C chemokine receptor type 5 (CCR5) gene encoding a human cell surface protein that has been implicated in HIV-1 entry. A naturally occurring 32-base-pair (bp) deletion (Δ32) in this gene in some human populations is believed to confer resistance for homozygous carriers to some HIV-1 strains, with a few other health- or immune-system-related implications7,8. Therefore, it has been proposed that inducing similar mutations in patients with HIV could be therapeutic. A clinical trial transplantation of autologous CD4+ T cells in which CCR5 was disrupted to HIV-infected individuals resulted in HIV rebound in all individuals who received the cells, likely due to low gene-editing efficiency9.
The main limitation of conventional gene editing is that there are only a handful of cases among thousands of pathogenic germline mutations for which inducing additional de novo mutations could be therapeutic. This concept is unlikely to meet the high efficacy and safety requirements for GGT because of the uncertainty associated with inducing novel gene mutations in children. Indeed, public response to a recent case of CCR5 disruption in human embryos that reportedly resulted in birth of twin girls in China10 was unanimously negative. The statement released by the organizing committee of the Second International Summit on Human Genome Editing in Hong Kong on 29 November 2018 summarized that “even if the modifications are verified, the procedure was irresponsible and failed to conform with international norms”10.
We will discuss below in more detail recently developed gene therapy strategies that are based on the replacement of mutated genes with normal wild-type copies (such as for mitochondrial DNA (mtDNA) mutations) or actual repair of endogenous mutant sequences and conversion of them back to normal using endogenous DNA-repair mechanisms. We believe these technologies meet the stringent safety and efficacy standards expected for GGT and are ethically acceptable since they do not induce new genetic modifications.
In this regard, we would like to note that GGT may utilize various genetic manipulation methods, including gene editing and gene replacement, to achieve the therapeutic goal, which is to modify genes such that they become the wild-type variants that are common in the human population; thus, in our opinion, if GGT is carried out such that it avoids off- and on-target mutations, it should provide no long-lasting negative alteration to the germline.
Commonly, targeted sequencing of the mutant locus is used to detect embryos carrying a genetic disorder, and frequently acquired chromosomal abnormalities are identified by array comparative genomic hybridization (aCGH) or fluorescence in situ hybridization (FISH) approaches. With the availability of whole-genome and whole-exome sequencing data, the scope of PGD has expanded to include rare genetic diseases, and the American Society of Reproductive Medicine (ASRM) has recently extended ethical justification to the use of PGD in patients carrying mutations that are associated with age-onset diseases13.
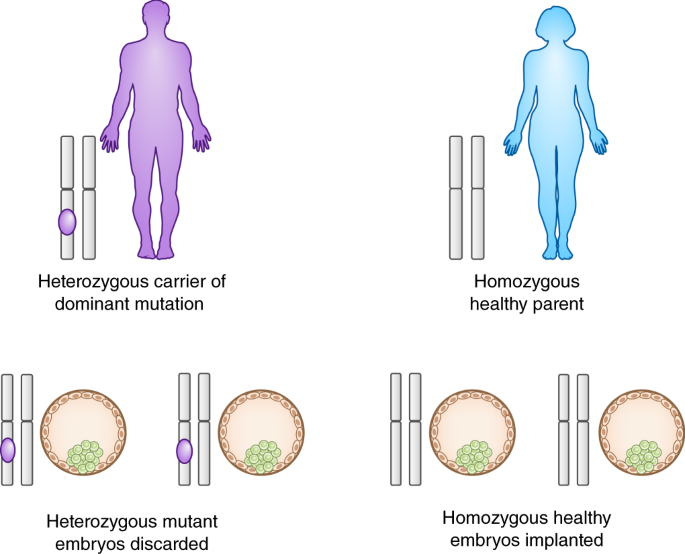
When only one parent in a couple carries a heterozygous mutation, 50% of the embryos produced by that couple on average would be mutation-free and available for transfer, while the remaining carrier embryos would not be (Fig. 1). However, according to the data collected by the Society for Assisted Reproductive Technology (SART), in 2016 the live birth rates following a single cycle of IVF, when all embryos are non-mutant, ranged from 3.3% to 47.6%, depending on the age of the women who went through the procedure14. This is attributed to poor embryonic development, spontaneous aneuploidy and low implantation rates of human embryos produced by IVF.
If one of the parents is heterozygous for a mutation, IVF success rates will likely be much lower, since a significant number of otherwise good-quality embryos will carry mutations and thus will not be selected for implantation. According to a recent study of 358 families at risk of transmitting a single-gene disorder who were undergoing IVF and PGD, the live birth rate was 18% compared to 38% for couples that underwent IVF for infertility (not genetic disease) during the same period15. This low IVF success rate was primarily contributed to by the fact that 959 embryos were carriers of gene mutations but otherwise were fit for transfer and thus were not implanted. Thus, couples with genetic mutations undergoing PGD selection experience significantly lower birth rates, forcing them to go through additional IVF cycles in order to have a healthy child, which is an expensive and invasive process.
If, as many believe, life begins at conception, the intentional destruction or abandonment of a mutant embryo is of ethical concern and is often an unacceptable option for many parents. In the United States, there are no federal limitations on the use of PGD, and while such decisions are left to the states, none have implemented laws with respect to PGD. The American Society for Reproductive Medicine has only published limited guidelines concerning PGD use.
In contrast to these selection-based approaches, gene repair in gametes or preimplantation embryos would rescue most or all mutant embryos, thereby increasing the number of embryos available for transfer and removing the need to dispose of a potential life. Therefore, GGT is more ethically acceptable to some families. Furthermore, GGT would provide an option to those families when PGD is not applicable, for instance, when one parent carries a homozygous autosomal dominant mutation or both partners are homozygous for the same autosomal recessive mutation, because all embryos would be affected.
GGT for mitochondrial DNA mutations
Maternally inherited mtDNA mutations give rise to a broad range of incurable conditions in children and adults. Those affected frequently die at an early age after an agonizing disease course. Moreover, prevention of transmission by way of PGD is compromised due to the uncertainty of the percentage of mutated mtDNA in the carrier’s oocytes16. The mitochondrial genome is resistant to direct DNA alterations made with gene-editing tools such as CRISPR due to the lack of native mtDNA repair and recombination mechanisms. However, an alternative form of GGT termed mitochondrial replacement therapy (MRT) has been developed that relies on the replacement of mutated mtDNA in the mutation-carrier’s oocytes with donated, mutation-free counterpart mtDNA, thereby allowing women carrying mtDNA mutations to circumvent passage of a condition to their children17. MRT is routinely accomplished via spindle transfer that is conducted at the mature oocyte stage when the nuclear DNA material is assembled into metaphase chromosomes forming a meiotic spindle. The spindle is microsurgically isolated into a karyoplast and then transplanted into the ‘empty’ cytoplasm of a donated unfertilized oocyte that itself has been enucleated. The reconstructed oocyte, now free of mutated mtDNA, can be fertilized and subsequently transferred into the patient18. Similar strategies involve the transfer of polar bodies or pronuclei19,20.
MRT technology is also relevant to the treatment of infertility, when a condition is secondary to age-related decline in oocyte cytoplasm quality17. Aging of the reproductive system in women of advanced age results in a drastic decline in the quality of oocytes, which subsequently leads to low pregnancy rates and a high percentage of pregnancy loss or birth defects. Recent evidence suggests that many factors responsible for oocyte aging are confined to the cytoplasm, and thus MRT, which is essentially whole cytoplasm replacement, may prove valuable for overcoming this form of reproductive aging.
When pioneered in a nonhuman primate, the MRT procedure resulted in the production of several live infants that possessed exclusively donor mtDNA18,21. We also demonstrated that the postnatal growth, development and reproductive capacity of these animals is normal and comparable to control animals produced by conventional IVF procedures22. The approach appears clinically relevant because preimplantation development of human MRT embryos was comparable to controls22,23,24.
While clinical trials involving the transfer of MRT embryos are not currently allowed in the United States where MRT was first developed, government-sanctioned first-in-human trials of MRT are presently underway in the United Kingdom25. MRT has been carried out already in Mexico, resulting in the birth of a healthy male child with donor mtDNA for a family carrying a deadly disease associated with mtDNA mutation26. This is the first successful case of GGT in humans that provides hope for the future of this therapy for families with heritable mtDNA gene mutations.
It is unfortunate though that in this case, affected families were forced to resort to a clinic located in a perhaps a less regulated jurisdiction, calling into question the regulatory utility of the moratorium on GGT in the United States27. Instead of an outright ban, perhaps a middle-of-the-road approach could be considered that involves clinical trials by selected academic centers with expertise to satisfy legitimate safety and efficacy concerns27.
The replacement of mutated mtDNA during the MRT procedure is not complete as a small fraction (1–4%) of maternal mtDNA remains in the oocyte or embryo. Such low levels of mutated mtDNA are not sufficient to cause a disease. However, its selective expansion during postimplantation embryonic and fetal development and rapid reversal back to the homoplasmic maternal mtDNA poses potential safety concerns28.
Recent studies revealed that zinc-finger nucleases (ZFNs) and transcription-activator-like effector nucleases (TALENs) can be used to target and selectively destroy the mutated mtDNA in mice29,30,31. These proof-of-principle findings suggest that programmable nucleases could be a therapeutic option for reduction of mtDNA mutation loads for heteroplasmic mtDNA diseases.
Correcting heterozygous mutations by gene conversion
In contrast to mtDNA, the nuclear genome is associated with robust DNA repair and recombination machineries that historically were used to experimentally induce a variety of gene modifications. Recent advances in genome-editing technologies allow induction of DNA double-strand breaks (DSBs) at specific loci in animals and human cells with high efficiency5. Current progress has been greatly facilitated by the development of DNA-cutting nucleases with programmable, site-specific DNA-binding domains, including RNA-guided CRISPR–Cas9 vectors5,32,33,34.
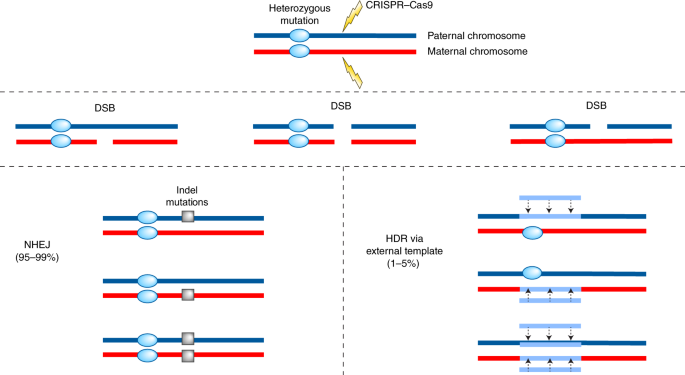
DSBs, along with other types of DNA damage, occur spontaneously as a result of environmental stress or DNA replication errors35. DSBs are typically repaired via non-homologous-end-joining (NHEJ) or homology-directed repair (HDR) mechanisms (Fig. 2). The emerging picture from recent genome-editing studies suggests that NHEJ is a prevailing repair pathway in somatic cells. However, this error-prone mechanism frequently involves introducing small indel mutations at the DSB site36. In rare cases, cutting both alleles simultaneously can cause large deletions or complex rearrangements, raising the possibility that neighboring genes or regulatory sequences could be affected37. As indicated above, NHEJ is the main expected outcome of gene editing and as such is highly undesirable for GGT applications as it produces additional mutations within or adjacent to the pre-existing germline mutation locus.
Both parental alleles carry the mutation and thus will be targeted by CRISPR–Cas9, resulting in DSBs on one or both alleles. Repair by NHEJ would dominate, producing additional indel mutations on one or both parental chromosomes. HDR via an endogenous ssODN template occurs less frequently, resulting in repair of the DSB and the preexisting germline mutation on one or both parental chromosomes.
Alternatively, cells can rebuild DSBs via HDR, a template-directed repair mechanism that takes advantage of existing homologous sequences in synthetic single–stranded oligodeoxynucleotide (ssODN) templates. In contrast to NHEJ, HDR ensures relatively accurate restoration of DSBs to the original sequence without gain or loss of DNA sequences36. More importantly, HDR extends further downstream and upstream from the DSB locus and not only repairs the break site, but also erases and rebuilds adjacent sequence variants that are different from the template. Thus, HDR can repair nearby pre-existing mutated sites, providing an opportunity to utilize this phenomenon to correct mutations. However, the frequency of HDR when both alleles are targeted (homozygous loci, Fig. 2) is substantially lower and ranges on average from 1–5%, while the majority of DSBs are resolved by NHEJ (95–99%)38,39. These outcomes, which could be acceptable for somatic gene therapy, are not sufficient for the more stringent safety and efficacy requirements raised for GGT applications.
Most efforts to increase HDR and lower NHEJ occurrence have focused on biasing repair outcomes by suppressing components of NHEJ while enhancing HDR pathways and manipulating the cell cycle38. These interventions that often employ chemical and genetic tools, although highly useful in research contexts in vitro, may be undesirable for therapeutic applications in human embryos because they may alter the capacity to respond to damage at other sites in the genome. Synthetic-template-mediated HDR frequencies can also be increased by optimization of design, orientation, polarity and length of the ssODN38.
Efforts toward GGT in human embryos are currently focused on utilization of the alternative endogenous sequences located on homologous chromosomes as a repair template using a mechanism known as gene conversion. Gene conversion is a unidirectional transfer of genetic material from a donor sequence to a highly homologous acceptor, and it does not require an exogenous template40. Gene conversion occurs during meiotic noncrossover recombination, is triggered by DSB repair and leads to the copying of intact homologous sequences to the region that contains the DSB. Gene conversion occurs in mitosis as well and may provide a strategy for correcting heterozygous mutations similar to known cases of spontaneous gene repair40. A current understanding of the mechanisms involved in gene conversion is reviewed in ref. 40.
We recently reported our study on the correction of a dominant germline mutation, a 4-bp deletion in the MYBPC3 gene that is associated with hypertrophic cardiomyopathy (HCM)41. We generated heterozygous mutant human embryos by fertilization of wild-type oocytes with sperm carrying the mutant copy of MYBPC3(MYBPC3∆GAGT). To induce a gene-conversion event, a single guide RNA (sgRNA)–Cas9 (refs. 42,43,44) construct was designed to bind and induce a DSB near the MYBPC3∆GAGT deletion, but not in the wild-type allele. We also synthesized an exogenous ssODN template encoding short homology arms to the target region (190 bp total length).
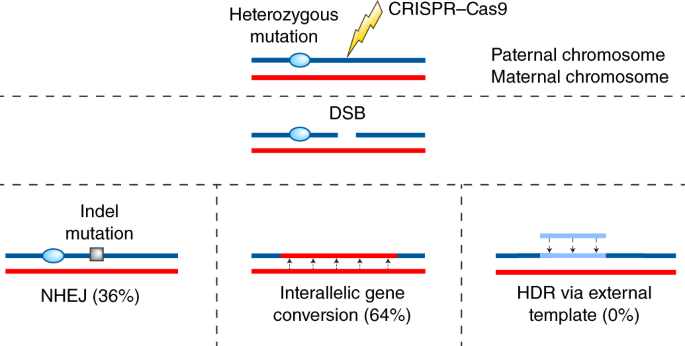
Using sequencing, we found that CRISPR–Cas9 induced DSBs at the mutated locus in heterozygous human embryos were preferentially repaired (64%) by gene conversion (Fig. 3). This resulted in the repair of the DSB and the MYBPC3∆GAGT loci in most of the human embryos, leading to a high yield of homozygous MYBPC3WT/WT embryos. Conversely, the incidence of NHEJ was significantly reduced (36%) compared to that in homozygous loci. Although ideally the incidence of NHEJ would be lower than this, even this efficacy indicates the correction of the mutation in the majority of mutant embryos, and thus fewer embryos have to be discarded than with conventional PGD. Interestingly, the exogenous ssODN included during the treatment was not used by the embryos.
CRISPR–Cas9 recognizes and induces DSBs on the mutant chromosome only, leaving the wild-type allele intact. DSBs are resolved via NHEJ but less frequently than seen for homozygous mutations. Gene conversion using the intact homologous chromosome occurs more frequently, resulting in repair of the DSBs and the germline mutation. HDR via an exogenous ssODN template is inhibited, possibly due to the failure to compete with the endogenous chromosome template. The percentages in the figure represent frequency at which each of the DNA repair mechanisms occurs in human embryos.
The exact mechanism and timing of mitotic gene conversion in human embryos remains to be investigated. However, our results challenge the prevailing opinion that interhomolog chromosome interactions and gene conversion are restricted to meiosis and are absent or very rare during mitotic cell divisions. Alternative interpretations of these results in humans have been considered and ruled out45,46,47.
Strategies for correcting nucleotide substitutions and homozygous mutations
Recent modification of gene-editing methods, termed base editing, allows conversion of one nucleotide bp to another. Direct bp conversion could become a viable alternative for the correction of both homozygous or heterozygous nucleotide substitutions. The main advantage of base editing over conventional gene editing is that it does not induce DSBs and thus circumvents NHEJ and the generation of indel mutations48. Base editing also does not require template-based HDR repair, the low efficiency of which is a major limitation for conventional gene editing. In contrast to HDR, base editing could be extended to quiescent cells due to the lack of requirement for cell proliferation. The procedure has been tested in cells grown in culture in a variety of species, including bacteria, plants and animals. Initially, the conversion of a C–G mutant pair to a wild-type T–A was described49. A more recent report on an A–T to G–C conversion employed deoxyadenosine deaminase50.
Existing base-editing approaches are also prone to off-target mutations; in particular, cytosine base editing was shown to induce a high frequency of unintended nucleotide changes in mouse embryos51. Overall, the limitation of base editing is its constraint to single-nucleotide substitution mutations and its relatively new application, meaning that there is limited experience with this technology.
Another limitation is that the mutant bp must be located at a certain distance from the nearby protospacer adjacent motif (PAM). A PAM is a short DNA sequence, typically 2–6 bp in length, downstream of the target region that is essential for CRISPR binding. This requirement limits the number of mutation sites in the human genome that can be efficiently targeted.
The interallelic gene conversion approach discussed above is unsuitable for correcting homozygous mutations. Hence, alternative strategies for correcting these mutations, such as increasing the incidence of HDR with exogenous templates, must be explored. As indicated above, in the study we carried out, the average HDR rate with ssODN for homozygous mutations was low. However, in meiosis, the rate of gene conversion is directly proportional to the length of the template sequence40. Therefore, it is likely that the length of commonly used ssODNs (<200 bp) is suboptimal. It is also possible that the specific design of the exogenous DNA template may promote HDR events. For example, use of double-stranded as opposed to single–stranded oligodeoxynucleotides.
Theoretically, GGT could be used to treat infertility caused by gene mutations affecting development of functional gametes or causing embryonic and fetal demise. For example, GGT could correct some genetic defects that would allow completion of the cycle of spermatogenesis. However, such therapeutic approaches will be complicated by the necessity of reliance on in vitro development and maturation of human gametes. Obviously, GGT interventions are more likely to be effective for treatment of preimplantation embryos, given that some forms of infertility are associated with fertilization failure or early embryonic arrest.
Safety Considerations
On- and off-target mutations
The introduction of unintended mutations or other genetic alterations in the course of CRISPR–Cas9-mediated correction of specific mutations in embryos could occur at genomic regions that are homologous to the targeted sequence, and the prevention of this must be addressed in preclinical studies52,53,54,55,56. The propensity to induce off-target DSBs may vary significantly depending on each targeted locus, the selected sgRNA and individual embryonic genome variations. Therefore, extensive screening for the optimal sgRNA using parental DNA, somatic cells and induced pluripotent stem cells (iPSCs) must be performed for each therapeutically targeted mutation before experimentation with human embryos. This step is particularly important when targeting heterozygous loci in which sequence differences between mutant and wild-type alleles can be minimal.
Potential off-target interactions can also be tested on somatic cells or iPSCs derived from the carrier parents. Typically, whole-genome sequencing, whole-exome sequencing or Digenome sequencing (an approach that specifically identifies off-target mutations) approaches are used to search for potential abnormalities57,58. DNA sequences from the CRISPR–Cas9-treated iPSCs from parents are then compared to sequence reads produced from untreated control DNA. Differences not attributable to sequencing errors may reflect potential off-target mutations. However, it should be noted that DNA profiles from potential parents are different from that in their embryos due to recombination and new heterozygosity.
Therefore, safety and efficacy results derived from parental samples should not be considered conclusive and must eventually be re-tested and reproduced in preimplantation embryos. This typically can be achieved by sequencing DNA samples derived from biopsied embryos treated with CRISPR–Cas9, similar to PGD. Due to the small sample size, DNA from embryos must be pre-amplified, introducing the potential for amplification errors and bias. In contrast to analysis of parent DNA samples, screening for off-target cleavage in embryos is also complicated because of the absence of untreated control DNA. DNA sequences from each embryo can be compared to DNA from parents and reference human DNA samples (representative example of human DNA), but potential off-target modifications must be differentiated from normal recombination sequence variations.
Another safety concern is that the synthesis of Cas9 protein from plasmids can lead to high enzyme concentrations and high levels of off-site targeting59. Fortunately, purified recombinant Cas9 protein (instead of plasmid) can be directly injected into human oocytes and zygotes, and that may enhance specificity while shortening enzymatic exposure time, thereby, diminishing off-site targeting41.
A critical safety concern that is often overlooked is undesirable on-target mutations, common for gene editing. As discussed, repair by NHEJ introduces additional indel mutations that could be more frequent and often more detrimental than off-target consequences. Besides small deletions or insertions generated by NHEJ, some DSB repair can involve large deletions extending up to several thousand bp in length37. In addition to large deletions, targeted alleles may also contain complex, often noncontiguous lesions, such as insertions of DNA segments from other chromosomes, inversions, duplications and single-nucleotide variations60. Therefore, it is desirable to reduce the frequency of NHEJ and to monitor on-target mutations using long-range PCR.
As outlined above, interallelic gene conversion extends beyond the DSB locus, resulting in the conversion of the neighboring mutant site and neutral parental SNPs and leading to loss of heterozygosity (LOH)45. Our recent results suggest that such gene conversion and LOH can expand considerable distances in both directions from the original target site (up to 10 thousand bp in length), and the length of the conversion tract can vary among individual blastomeres even from the same embryo. While conversion of the mutant locus to the wild type is desirable, extensive LOH is detrimental as it could also copy silent mutations from one parental allele into another. Such homozygous mutations, if functional, could have detrimental consequences. Sequence analysis of rat offspring produced by gene conversion revealed that the conversion tract length varied between 2–30 thousand bp around the targeted site61.
Recent studies have indicated that DSBs incite a p53-mediated DNA damage response and subsequent cell cycle arrest and apoptosis. Incidentally, gene editing can select for rare populations of cells with p53 mutations, leading to tumor formation62,63.
Mosaicism
A common issue that arises during GGT in embryos is mosaicism, the presence of unrepaired (mutant) and repaired (either or both NHEJ and HDR) cells within a multicellular targeted embryo or offspring64,65,66. Mosaicism complicates the PGD screening process as well, in which 3–5 cells are analyzed and extrapolated to the whole, and decisions are made on the basis of this. While in most cases CRISPR–Cas9 is injected into a single-cell embryo (a zygote), the actual repair is likely delayed and occurs after embryo cleavage, at the 2- or 4-cell stage. Strategies to reduce mosaicism involve shortening the half-life of Cas9 activity66,introducing CRISPR–Cas9 into early-stage zygotes67, or, as we demonstrated, injection of CRISPR–Cas9 into M-phase oocytes at the time of sperm introduction41. The latter can significantly reduce or completely abolish mosaicism in the case of a heterozygous mutation. Such early delivery likely allows degradation of CRISPR–Cas9 components before the first mitotic division.
Another approach to reduce mosaicism is increasing gene conversion efficacy while reducing repair by NHEJ. This is achieved by the co-injection of CRISPR–Cas9 with sperm into M-phase oocytes. We observed a targeting efficacy of 100% for injection into oocytes compared to 72% when CRISPR–Cas9 was injected into zygotes. Interallelic gene conversion rates were also higher in the M-phase injected group, resulting in an increased percentage of non-mosaic wild-type embryos (72.4%), compared to 66.7% for zygote-injected groups or 50% for untreated controls. In addition, repair rates can be enhanced by treatment with the gene conversion pathway member RAD51. Recent studies have indicated that exposure of embryos to RAD51 simultaneously reduces NHEJ while boosting gene conversion rates by threefold68,69.
Ethics and regulations
Ethical and regulatory issues surrounding clinical applications of human GGT have been widely reviewed and debated by several bodies, including an interdisciplinary ethics consortium called the Hinxton Group70, the US National Academies of Science, Engineering, and Medicine (NASEM)71, the American Society of Human Genetics (ASHG) Workgroup on Human Germline Genome Editing72 and most recently by the UK Nuffield Council on Bioethics73.
The consensus is that research on human preimplantation embryos must be allowed and should be carried out to evaluate safety, feasibility and efficacy and to establish standards for future clinical use, but only with appropriate oversight and consent. In this regard, the ASHG has strongly urged that public funding be provided for research on human preimplantation embryos and fetuses and warned that the continued ban on public funding could lead to behind-the-scenes rogue experimentation72.
Recommendations from all of these bodies support future clinical trials of human germline genome editing, but only for compelling medical needs, with credible preclinical and clinical evidence on risks and potential health benefits and subject to comprehensive oversight protecting the research subjects and their descendants. NASEM recommended a set of criteria that must be met for initiating clinical trials, including absence of reasonable alternatives, restriction to prevention of a serious disease and restriction to conversion of such genes to versions that are prevalent in the population and are known to be associated with ordinary health with little or no evidence of adverse effects.
In the wake of recent controversy surrounding the birth of gene-edited babies in China, which did not adhere to these recommendations and research ethics, a recent commentary proposed a global moratorium on clinical applications of human germline editing74. While this instant reaction is understandable, more than 30 countries including China already have regulations and laws in place prohibiting genetic modifications to the human germline. In the United States, the National Institutes of Health (NIH) has long been prohibited from funding any human embryo research. As mentioned above, since December 2015, the US Congress has included provisions in annual federal appropriations laws that prohibit the Food and Drug Administration from considering any application to create children from embryos that have been genetically modified. Thus, a pressing issue is not an additional moratorium or bans, but how to reinforce already existing regulations around the world.
Some view PGD as a reasonable alternative and thus suggest that GGT be limited to a few cases when PGD cannot be applied75,76. Such cases could include when one parent carries a homozygous autosomal dominant mutation, or when both parents are homozygous for the same autosomal recessive mutation. However, such scenarios are exceptionally rare in reality due to early disease onset and the severity of clinical manifestations that often lead to premature death.
The report by the Nuffield Council on Bioethics found no clear distinction between disease prevention and enhancement. GGT aims to correct genetic defects, while genetic enhancements seek to modify the genes that enhance the capabilities beyond what is normal. The council recommended legal changes in the United Kingdom to permit heritable genome-editing interventions that avoid inherited disease or enhance other characteristics. However, such interventions “must be intended to secure, and be consistent with, the welfare of the future person, and they should not increase disadvantage, discrimination or division in society”.
Changes in public attitudes toward somatic gene therapy and GGT were recently surveyed among 1,600 US adults from whom data was collected by YouGov between December 2016 and January 2017 (ref. 77). In contrast to previous polls, the majority of people found that both somatic gene therapy (64%) and GGT (65%) are acceptable. However, while 59% of respondents supported human genome editing to treat disease or to restore health, only 33% expressed at least some support for enhancement or improvement of human abilities. Another study reported similar results, suggesting that the majority of people support both somatic and germline gene therapy, but are far less enthusiastic toward enhancement78.
Currently, the United Kingdom is the only country in the Western world to give license to its researchers to conduct clinical trials involving germline gene therapy. The license is limited to mitochondrial-replacement therapy (MRT), wherein inheritance of mtDNA-based disease is circumvented by replacement of mutated mtDNA with donor mtDNA.
Clinical applications of GGT most likely will require additional preimplantation and/or prenatal biopsy and diagnostics to confirm accurate on-target repair but eliminate embryos with undesirable outcomes. It will be necessary to follow up on children born after GGT to monitor their development and health. There is a debate on how long children must be monitored, with opinions ranging from a few year years to decades, or even across generations79.
Future directions
With encouraging preliminary results in gene correction in mice and human preimplantation embryos, it may be time to focus on the enhancement of the efficacy of GGT and its extension to heritable pathogenic mutations in the human germline. Strategies that limit Cas9 activity or impact DNA repair machinery might be useful to explore, such as RAD51 on enhancing HDR as demonstrated in mice. Assessment of the biological risks of designer nuclease-mediated gene correction should also be further examined in model animals and preimplantation-stage human embryos.
(원문: 여기를 클릭하세요~)