Bacterial communication is a potential strategy to control bacterial behaviours and thus, attenuate pathogen infectivity; however, identifying the signalling molecules that regulate communication pathways is challenging. Now, a robust strategy to rapidly identify previously unknown signalling peptides has been developed. This approach provides a means to map out and decipher bacterial signalling mechanisms.
Communication pathways facilitate the ability of bacteria to work together when in sufficient numbers and can influence their pathogenicity as well as their ability to acquire resistance to existing antibiotics. Understanding communication pathways can thus enable the development of treatments that reduce the risk of resistance development. However, each species utilizes a unique signalling molecule for communication with only limited cross-talk between species. This diversity in signalling requires that each molecule be identified and characterized for every species being considered. Now, a team led by Christian A. Olsen report1 a method that greatly streamlines the identification of unknown signalling molecules by providing a simple, yet powerful strategy of isolating and characterizing previously unknown thiolactone-containing signal peptides.
The accessory gene regulator (agr) bacterial communication system is common in Gram-positive bacteria2,3,4,5. This system is used to facilitate group behaviours among populations of bacteria upon reaching a certain threshold, often called quorum sensing. To assess cell density and trigger group behaviours, the agr communication system commonly utilizes a macrocyclic peptide as the signalling molecule, which can only activate a membrane-bound receptor at sufficiently high concentrations in the media. This peptide is first generated in the cell as a longer, linear propeptide before being processed by cellular machinery into the shorter, cyclized final peptide that is exported out of the cell. Upon activation of the receptor by the peptide, a signalling cascade occurs, resulting in upregulation of genes involved in group-beneficial phenotypes. This includes the genes involved in producing, processing and detecting the signalling peptide (autoinduction). The group-beneficial phenotypes that are under quorum sensing control include swarming, luminescence, competence, biofilm formation and production of virulence factors6,7. Many of these group behaviours enhance the pathogenicity of the bacteria, making quorum sensing a potential therapeutic target.
Structurally, the macrocyclic peptides are typically either lactones or thiolactones, with the macrocyclic linkage made between the C-terminus of the processed peptide and an internal serine or cysteine, respectively2,3,4,5. Although these peptides have similarities, it is rare for a peptide to be able to activate the quorum sensing circuit of another species. More often, foreign signalling peptides can inhibit other species’ quorum sensing by binding to the receptor, but not activating it, while the most common result is that there are no significant cross-interactions at all. Because of the species-specific response to the peptide signals, it is typically necessary to identify and characterize the target peptide for each species. However, the identification is complicated by the fact that the propeptide undergoes processing before the mature peptide is produced. This means that the mature peptide cannot be directly predicted from genomic data and must be isolated from bacterial cultures. Moreover, the peptide is often present in comparatively low concentrations and undergoes relatively rapid degradation. This limitation has slowed and restricted the ability of researchers to accurately identify these signalling peptides in new species.
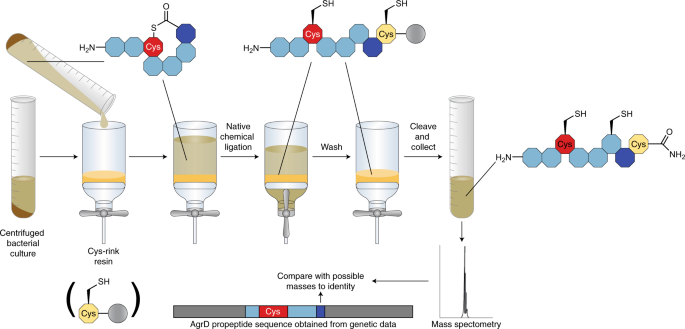
Olsen and co-workers have now developed a methodology that addresses these limitations for thiolactone peptides1. Their strategy takes advantage of the well-developed chemistry of native chemical ligation8 to selectively extract thiolactones from the supernatants of bacterial cultures and trap them on a solid phase resin as a linearized peptide with an additional C-terminal amidated cysteine. Native chemical ligation is a chemoselective reaction used to couple unprotected peptide fragments into large proteins. It involves a series of reversible thiol-thioester exchange reactions that terminate once an N–terminal cysteine is reached through an S to N acyl shift that affords a native amide bond, leaving all internally located cysteines intact. In the peptide-trapping methodology, the cysteine that is first attached to the resin reacts with the thioester bridge, opening the macrocyclic ring in the process of trapping the target peptide. Washing and cleavage from the resin allows analysis of the enriched peptides via mass spectrometry. Combining the mass spectrometry information with knowledge of the gene that encodes the matching peptide sequence enables rapid identification of the mature signalling peptide (Fig. 1). Olsen and co-workers have vetted their approach by successfully capturing and identifying five known Staphylococcus peptides before they set out to identify 11 unknown Staphylococcus peptides (identifying a total of 16 signalling peptides). The newly identified peptides were also re-synthesized using a known on-resin cyclisation–cleavage approach and subsequently tested for their potential as quorum sensing modulators. These assays showed a variety of potencies and two peptides were discovered to be inter-species agonists. Overall, the presented strategy allows for greatly accelerated identification of unknown peptides with high fidelity.
The supernatant of centrifuged bacterial culture contains the macrocyclic peptide. Mixing the supernatant with Cys-loaded rink amide resin along with native chemical ligation reagents results in trapping of the linearized peptide with its C-terminus attached to the cysteine on the resin. Once trapped, the resin can then be washed to remove contaminants and the peptide cleaved from the resin under acidic conditions, giving the linear peptide with an additional C-terminal amidated cysteine. This peptide can be analysed via mass spectrometry and identified by comparing to possible masses generated using the genetic sequence of the propeptide, AgrD. Figure adapted from ref. 1, Springer Nature Ltd.
The strength of the methodology developed by Olsen and co-workers lies in its simplicity, which makes it readily accessible to a wide range of researchers. All that is needed is the ability to culture bacteria, some relatively inexpensive reagents, a mass spectrometer and genomic data on the propeptide. Although this method only works with thiolactones, the diversity of Gram-positive genera that utilize or are predicted to generate such signalling molecules makes this methodology widely applicable. The method’s chemoselectivity also reduces the risk of erroneous side reactions. Moreover, the sensitivity of the method is more than adequate for the questions in hand since peptide supernatant concentrations as low as 12.5 nM can be detected while thus far the concentrations of these macrocyclic peptides were found to be in the low micromolar range in bacterial supernatants.
A possible limitation of this methodology is the potential presence of competing side reactions that might give some ambiguity in results, if they were not anticipated. For example, there is the possibility of a S to N acyl shift for thiolactones formed between the C- and N-termini, which could render a peptide undetectable by this method. In addition, the potential formation of a disulfide bridge in the product peptide instead of the normally obtained linear peptide needs to be kept in mind. When dealing with some of these cases, the timing of peptide extraction from the cell cultures and the specific growth conditions could have a large impact on whether the peptide is successfully detected.
While only a handful of peptides had been identified and characterized in the last few decades, the method has already enabled the team to more than double that number during their study. Adopting this method will allow for the rapid identification of further unknown signalling peptides in an array of Gram-positive bacteria, particularly any that have an agr-like quorum sensing circuit. We expect this method to be transformative by characterizing communication pathways between populations of bacterial species. The approach will also aid in the identification of new potentially therapeutic lead compounds that target quorum-sensing-dependent infections. The largest challenges for identifying unknown thiolactone peptides that remain are difficult-to-culture bacteria and obtaining the genomic information for lesser-known bacteria. With the development of this method, the isolation step will no longer be an obstacle for this subclass of signalling molecules.
(원문: 여기를 클릭하세요~)