(원문)
Chromosomes can exist outside the nucleus in rupture-prone structures called micronuclei. It emerges that micronuclei are fragile because their outer layer lacks some nuclear-envelope components.
The genome sequencing of some cancer cells revealed the occurrence of large-scale DNA rearrangements that are characteristic of a single catastrophic event1. This type of genetic abnormality is termed chromothripsis. It occurs in many types of human cancer, and is thought to drive tumour formation2. Chromothripsis can arise as a result of cell division if individual chromosomes do not join the other chromosomes within the newly formed nuclei but instead form rupture-prone structures called micronuclei3,4. Writing in Nature, Liu et al.5 report that the nuclear-envelope structure that encloses a micronucleus lacks some of the components of the nuclear envelope that surrounds a nucleus. They propose a mechanism for how this might occur, and how it might account for micronuclear fragility.
In the cells of organisms such as animals and plants, the genome is stored in a nucleus. This is surrounded by a nuclear envelope6 composed of lipid membranes, proteins, and multi-protein structures termed nuclear-pore complexes, which provide a route for the transport of materials between the nucleus and the cytoplasm. During cell division, the nuclear envelope disassembles, and the replicated chromosomes bind to a structure called the spindle, which is made of protein filaments called microtubules. The chromosomes align on the spindle at the centre of the cell, and the two copies of each chromosome then separate and move away from the middle of the cell. At a later stage of cell division termed telophase, a nuclear envelope reassembles around the chromosomes to form a nucleus in each daughter cell.
However, if any chromosomes lag behind the rest and are still in the middle of the dividing cell during telophase, they become isolated and can form a separate small nucleus called a micronucleus. Micronuclei often rupture spontaneously7, resulting in substantial damage to DNA and chromothripsis3,4. The basis for micronuclear fragility was unclear.
Studying cell division in human cells grown in vitro, Liu et al. found that lagging chromosomes formed micronuclei that lacked certain components of the nuclear envelope, such as proteins from the nuclear-pore complex and proteins called B-type lamins that are necessary for nuclear-envelope integrity. Indeed, deficiencies in B-type lamins were previously shown to trigger micronuclear fragility7. Deficiencies in the formation of nuclear-pore complexes in the nuclear envelope that surrounds a micronucleus might hinder the entry of the proteins needed for successful repair of DNA damage and for stabilization of the nuclear envelope, which might explain why micronuclei are prone to rupture7 and chromothripsis3,4.
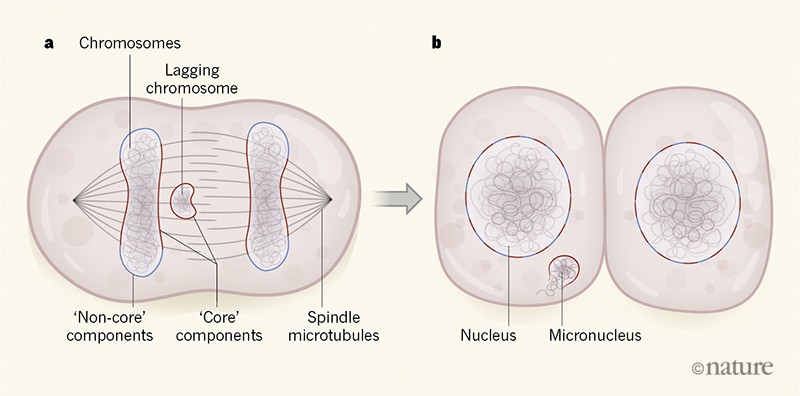
When normal nuclear assembly occurs around the end of cell division, part of the emerging nucleus is in close contact with the spindle, where the local density of microtubules is high. This part of the emerging nucleus is referred to as a region that contains ‘core’ nuclear-envelope components (Fig. 1). The part of the nucleus that emerges in areas with low microtubule density is known as the region that contains ‘non-core’ nuclear-envelope components. The authors found that nuclear-envelope components that incorporated efficiently into micronuclei were those components known8 to localize to the core regions of reassembling nuclei. The nuclear-envelope components that were inefficiently incorporated or absent from micronuclei were those that are found on the non-core regions of reassembling nuclei.
Figure 1 | The formation of a micronucleus. During cell division, chromosomes align in the middle of the cell on the spindle, which is made of protein filaments called microtubules. a, When chromosomes then move away from the middle of the dividing cell, a lagging chromosome might remain stranded in the centre of the cell, owing to a cell-division error. As cell division nears completion, the chromosomes become surrounded by what is termed nuclear-envelope material. ‘Core’ components of the nuclear-envelope material reassemble around chromosomes in spindle regions that have a high density of microtubules, and ‘non-core’ components reassemble in regions with a low density of microtubules. Liu et al.5 analysed cell division in human cells grown in vitro. They report that the high density of microtubules surrounding lagging chromosomes prevents the binding of non-core components of the nuclear envelope.b, When nuclear-envelope assembly is completed, the core and non-core materials become interspersed in the nuclear envelope. When nuclear-envelope material reassembles around a lagging chromosome, the result is a rupture-prone structure termed a micronucleus that can undergo large-scale DNA damage and is linked to cancer2–4.
Liu and colleagues investigated whether the failure of lagging chromosomes to recruit non-core nuclear-envelope components depends on the presence of the spindle. The authors treated cells undergoing cell division with drugs that dismantled the spindle. This treatment enabled both core and non-core nuclear-envelope components to assemble on lagging chromosomes that were forming micronuclei. The authors also conducted experiments in which they manipulated cells to displace some chromosomes away from the spindle to peripheral regions of a dividing cell. These displaced chromosomes formed micronuclei that had both core and non-core nuclear-envelope components. Such micronuclei did not rupture, revealing that non-core components are needed to prevent micronuclear fragility. The authors’ results are consistent with a model in which the micronuclear assembly of nuclear-envelope material is perturbed in regions of the cell that have a high density of microtubules.
Cell division is a highly regulated process, and there are mechanisms to ensure its quality control. This raises the question of whether any such checkpoints exist in human cells to deal with lagging chromosomes.
In the fruit fly Drosophila melanogaster, there is a gradient of activity of the kinase enzyme Aurora B that is highest near the spindle in the centre of a dividing cell9. This enzymatic activity inhibits nuclear-envelope reassembly on lagging chromosomes that are within the region of high Aurora B activity. This mechanism for inhibiting nuclear-envelope reassembly might facilitate the reintegration of a lagging chromosome with the rest of the chromosomes that will form the new nucleus9. Indeed, when Aurora B is mislocalized away from the spindle in Drosophila cells9, nuclear-envelope material, such as the non-core, nuclear-pore-complex protein Nup107, binds to lagging chromosomes with the same kinetics as to the main chromosome group.
However, when Liu et al. caused similar perturbations of Aurora B in human cells, this did not restore the recruitment of a non-core, nuclear-pore-complex protein (in this case, Nup133) to lagging chromosomes. The contrasting outcomes might reflect species-specific differences in sensitivity to Aurora B perturbations. Liu and colleagues found that core nuclear-envelope components accumulated on lagging chromosomes with normal kinetics and that drug-mediated inhibition of Aurora B at a late stage of cell division just before nuclear-envelope reassembly did not result in the recruitment of non-core, nuclear-envelope components to lagging chromosomes. These findings suggest that a dedicated chromosome-separation checkpoint is unlikely to exist in human cells. Instead, chromosome segregation to the daughter cells and nuclear-envelope assembly are not closely coordinated events. This could explain why lagging chromosomes often form micronuclei with deficient nuclear envelopes, rather than being incorporated into the nucleus.
How microtubules might limit the access of non-core, nuclear-envelope components to lagging chromosomes is unknown. The non-core and core components of the nuclear envelope both include membrane-bound proteins that redistribute during cell division to an organelle called the endoplasmic reticulum. One possibility is that these core and non-core proteins localize to different parts of the endoplasmic reticulum. Perhaps sheet-like domains of the organelle that are too large to enter spindle regions that have high microtubule density might contain non-core proteins, whereas the smaller tubular structures of the endoplasmic reticulum that might be able to enter the spindle regions could contain the core proteins. Alternatively, perhaps the localization of core and non-core nuclear-envelope proteins might be regulated by the microtubule-associated proteins that remove nuclear-envelope lipid membranes from chromosomes during an early stage of cell division10. It will be exciting to learn the outcome of studies to test these and other possible mechanisms, because the results might provide insights into general principles of how microtubules and other filament-like networks in the cell regulate the membrane-bound outer layer of organelles.