(원문)
An investigation finds that most protein complexes in yeast cells assemble before the subunits have fully formed. This mechanism might prevent the formation of toxic protein aggregates.
Most cellular processes are carried out by proteins, which generally assemble into heteromeric complexes — those composed of two or more distinct subunits. Although it was thought for many years that protein subunits diffuse freely in the cell and form complexes through random collisions, this seems unlikely, given that the cellular environment is extremely crowded. Writing in Nature, Shiber et al.1provide in vivo evidence that, in eukaryotic organisms (which include animals, plants and fungi), most protein complexes in the cytoplasm are assembled co-translationally — that is, assembly occurs while at least one of the subunits is still being synthesized by the cell’s ribosome machinery.
The study of co-translational protein-complex formation in vivo was challenging until a technique known as ribosome profiling was developed2 in 2009. This technique allows the positions of ribosomes on messenger RNAs to be determined by sequencing RNA fragments, and is usually used to monitor translation — the process in which the ribosome decodes mRNA and uses it as a template for protein synthesis. Shiber et al. used a modified protocol called selective ribosome profiling3, which isolates ribosomes that are synthesizing nascent protein chains already interacting with another protein. Subsequent sequencing of the corresponding RNA fragments reveals the mRNAs that encode the interacting nascent chains. The sequencing also identifies the protein domains involved in the interaction, because only ribosomes bound to nascent chains that contain fully exposed interaction domains will be isolated by selective ribosome profiling.
The authors investigated the prevalence of co-translational protein-complex assembly for 12 stable, well-characterized heteromeric complexes in the yeast Saccharomyces cerevisiae. They found that 9 of the 12 complexes assembled in a co-translational manner. Although it had been suspected that some protein complexes are assembled co-translationally4,5, the finding that this process is widespread is surprising. Notably, the three protein complexes that did not seem to do this use dedicated chaperone proteins to assist in assembly. Given that the major function of chaperones is to prevent misfolding and random aggregation of proteins during protein folding6, the researchers hypothesized that co-translational protein-complex assembly might serve a similar purpose.
Shiber et al. went on to find that six of the nine complexes that assemble co-translationally have a directional assembly mode: one of the subunits must be fully folded before it engages the nascent chain of a second subunit, but the fully folded second subunit cannot engage the nascent chain of the first subunit. This means that the second subunit must always participate in co-translational assembly as a nascent chain. Intriguingly, when the authors studied yeast strains that had been engineered not to produce the fully folded subunit, they observed that the nascent chain of the second subunit forms aggregates. This indicates that co-translational assembly does indeed prevent the formation of potentially toxic protein aggregates.
Although the authors convincingly show that co-translational protein-complex assembly is widespread, it is unclear how the subunits are brought into proximity to enable complex formation. There are two plausible, broad models, which are not mutually exclusive.
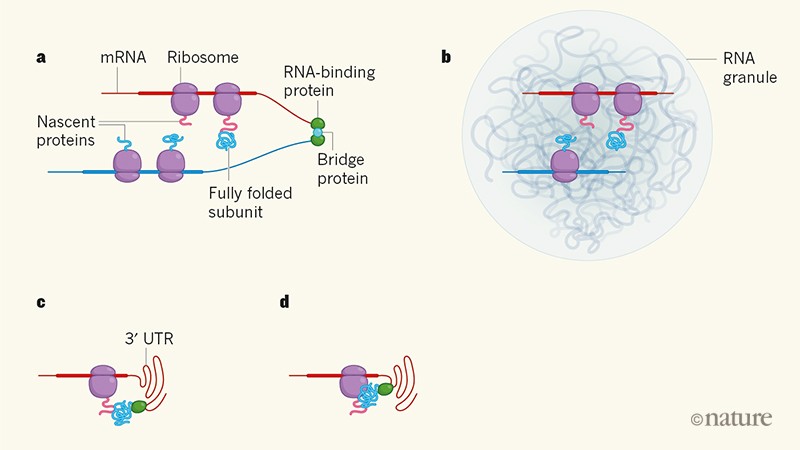
In the first model, the two mRNAs that encode the subunits are translated near to each other. This proximity could be achieved by an RNA-binding protein that bridges the mRNAs (Fig. 1a). Such a scenario has been suggested for the assembly of heteromeric ion channels7. However, a genome-wide analysis of human RNA–protein interactions8found that it is rare for two different mRNAs to be bridged by an RNA-binding protein. But a physical linkage is not necessarily needed to bring two mRNAs close together: mRNAs that share certain sequence motifs could co-localize in RNA bodies known as granules (Fig. 1b), thereby allowing them to be translated at defined subcellular locations9.
Figure 1 | Possible ways in which protein subunits are brought together for co-translational assembly. The cell’s ribosome apparatus uses the sequences of messenger RNAs as templates for protein synthesis. Shiber et al.1 report that the subunits of most cytoplasmic protein complexes in the yeastSaccharomyces cerevisiae are assembled co-translationally (that is, while at least one of the subunits is still being synthesized). But how are the different protein subunits brought together for assembly into complexes? a, One possibility is that an RNA-binding protein bridges two mRNAs, either directly (not shown) or indirectly (shown) through another protein. Here, a recently fully folded subunit formed on one mRNA has detached from its ribosome (which has disassembled; not shown), and interacts with a nascent protein on another mRNA. Thick regions of mRNAs represent translated regions; thin regions are not translated. b, The mRNAs could also come into close proximity in bodies known as RNA granules. c, An untranslated region of mRNAs known as the 3ʹ untranslated region (3ʹ UTR) might also recruit a fully folded subunit using an RNA-binding protein, bringing the subunit close to the mRNA’s nascent protein. d, In a variant of c, the recruited subunit is temporarily deposited on the ribosome before being transferred to the nascent protein.
In the second model, the two mRNAs are not close to each other. Instead, an RNA-binding protein recruits a fully folded protein subunit to an untranslated region (the 3ʹ UTR) of an mRNA that encodes the second subunit, thus allowing the folded subunit to interact with the second subunit as the latter is synthesized. In mammalian cells, 3ʹ UTRs of mRNAs have been shown to recruit proteins that then interact with the mRNAs’ nascent or newly made proteins10 (Fig. 1c). And in budding yeast, proteins recruited to mRNAs undergoing translation have been observed to be temporarily deposited on the ribosome before being transferred to the nascent protein chains (Fig. 1d)11.
Recruitment of interacting proteins by 3ʹ UTRs is reminiscent of co-translational protein-complex formation in bacteria, in which protein subunits are often encoded by a cluster of neighbouring genes (an operon) that are expressed as a group12,13. However, in bacteria, the only fully folded protein that can interact with nascent proteins is one of the encoded subunits, whereas the use of the 3ʹ UTR in eukaryotes might allow several different interactors to be recruited to nascent proteins — thus enabling a variety of protein complexes to be assembled co-translationally14.
More experiments are needed to work out how interacting protein subunits are brought together and how mis-assembly of complexes is prevented. Nevertheless, Shiber et al. have demonstrated that protein-complex formation often relies on recruitment mechanisms, rather than diffusion, to achieve specific protein interactions. Their findings add to an increasing number of in vivo observations suggesting that most cellular processes are interconnected: mRNAs not only encode proteins, but also increase the specificity of protein-complex formation by assisting the compartmentalization of proteins in the cytoplasm, and by regulating localized translation. Finally, it remains to be seen whether the majority of stable protein complexes in mammalian cells are also assembled co-translationally — but it seems likely that they are.